Abstract
For many familiar with metazoan relationships and body plans, the hypothesis of a sister group relationship between Diploblasta and Bilateria1 comes as a surprise. One of the consequences of this hypothesis - the independent evolution of a nervous system in Coelenterata and Bilateria - seems highly unlikely to many. However, to a small number of scientists working on Metazoa, the parallel evolution of the nervous system is not surprising at all and rather a confirmation of old morphological and new genetic knowledge.2-4 The controversial hypothesis that the Diploblasta and Bilateria are sister taxa is, therefore, tantamount to reconciling the parallel evolution of the nervous system in Coelenterata and Bilateria. In this addendum to Schierwater et al.,1 we discuss two aspects critical to the controversy. First we discuss the strength of the inference of the proposed sister relationship of Diploblasta and Bilateria and second we discuss the implications for the evolution of nerve cells and nervous systems.
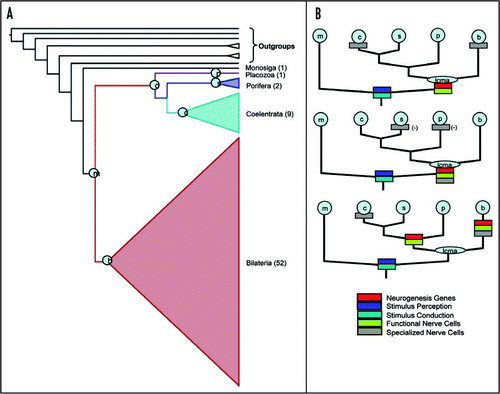
The analysis in Schierwater et al.Citation1 involved 24 ingroup taxa and several carefully chosen outgroups. Here we present a larger analysis of 72 taxaCitation5 to reinforce the inference we obtained with the smaller taxonomic sample. presents the results of this analysis and shows clearly that the Bilateria and Diploblasta are monophyletic and sister to each other with robust bootstrap support for both parsimony and maximum likelihood analyses. We could not overturn the sister group relationship of these two groups regardless of the larger taxonomic sampling or the statistical tests we used in the present analysis (). It is clear to us from analyses with broader taxonomic representation that the sister relationship of Bilateria and Diploblasta is a valid hypothesis.
With respect to the controversial aspect of parallel nervous system evolution, we point out that a definition of a nervous system that satisfies most is that nervous systems are spatially organized systems of aggregated nerve cells. The simple question, “what is a nerve cell?” then becomes the crux of the argument. But, this question elicits a spectrum of answers from different experts. Accurate homology statements concerning nerve cells are crucial to the story and these have to wait for a general definition of what a nerve cell is. The key to these definitions lies in examining the non-bilaterian animals.Citation2,Citation6 In most modern views “early nervous system evolution” is the equivalent of “early co-evolution of electrical excitability and functional synapses organizing intracellular and extracellular signaling processes spatio-temporally”.Citation6 Most zoologists agree that neither Placozoa nor Porifera have nerve cells or a nervous system, but it is important to recognize that both sponges and placozoans show behavior. They respond in a coordinated way to external stimuli that must be perceived and mediated by some kind of perception and transduction cells. Both sponges and placozoans harbor a pre-nervous integration system with many so-called “nerve cell typical” features, molecules and related genes, but these characteristics cannot be co-localized with any specific cell type.Citation7–Citation10 While in sponges several cell types are likely involved in signal perception and transduction, in placozoans it seems to be a single cell type only, the fiber cells, which form a loose connection network in the center of the placozoan body.Citation11
Although we are far away from a general definition of a nerve cell (and therefore a definition for nervous system), we can still summarize our current knowledge on early nerve cell evolution () as follows: The last common metazoan ancestor (LCMA) likely possessed a pre-nervous system with some kind of unspecialized proto-nerve cells. Placozoa and Porifera cum grano salis conserved this stage, while both Coelenterata and Bilateria developed specialized nerve cells from this stage (top; scenario in ). In this light the parallel invention of nerve cells, and consequently a nervous system, in Bilateria and Coelenterata is hardly problematic and not much more than a morphological and physiological specialization of already existing proto-nerve cells. Since specialization of totipotent cells into unipotent cells is a routine step in all metazoan lineages it seems possible to evolve specialized nerve cells directly from proto-nerve cells. In other words, the invention of so-called nerve cells is anything but a major invention in metazoans, if the LCMA already possessed protonerve cells, which obviously seems to be the case.
Figures and Tables
Figure 1 (A) Phyologenetic tree with relationships within Bilateria, Coelenterata and Porifera collapsed. The 72 taxa are comprised of the 64 taxa from (5) plus eight taxa added from (1). Numbers in parentheses refer to number of species in each of these groups. Phylogenetic matrices and tree topologies within the collapsed groups are available from the authors. We inferred the phylogeny using a maximum likelihood (ML) and maximum parsimony (MP) optimality criterion. Node support values (ML/MP) for nodes marked by circles with inset letters are: (B) Bilateria 100/100, (C) Coelenterata 100/82, (S) Porifera 100/100, (D) Diploblasta 100/99, (M) Metazoa 100/63; (P) Placozoa is a single taxon. Within the Bilateria: Deuterostomia 100/100, Protostomia 100/100. (B) Phylogenetic scenarios for the evolution of nerve cells mapped onto the Diploblast-Bilateria Sister hypothesis. Five potential characters (represented by colored boxes in the figure) important in the evolution of nerve cells are mapped onto the Diploblast-Bilateria Sister. Most qualities of a nerve cell seem to have been present already in the last common metazoan ancestor (LCMA in light blue). In the top figure we present the most parsimonious explanation for the evolution of these five characters (6 parsimony steps). Only the specialization of multifunctional proto-nerve cells into unifunctional nerve cells would have occurred in parallel in Bilateria and Coelenterata in the above scenario. The middle scenario is similar to the top only instead of hypothesizing independent gain of specialized nerve cells it hypothesizes independent loss of specialized nerve cells (7 steps). The bottom tree shows a highly unlikely scenario where the number of steps is nearly twice that of the top scenario.
Addendum to:
References
- Schierwater B, Eitel M, Jakob W, Osigus HJ, Hadrys H, Dellaporta SL, et al. Concatenated analysis sheds light on early metazoan evolution and fuels a modern “urmetazoon” hypothesis. PLoS Biol 2009; 7:1000020
- Blackstone NW. A new look at some old animals. PLoS Biol 2009; 7:7
- Hanström B. Vergleichende Anatomie des Nervensystems der Wirbellosen Tiere 1928; Berlin Springer
- Srivastava M, Begovic E, Chapman J, Putnam NH, Hellsten U, Kawashima T, et al. The Trichoplax genome and the nature of placozoans. Nature 2008; 454:955 - 960
- Dunn CW, Hejnol A, Matus DQ, Pang K, Browne WE, Smith SA, et al. Broad phylogenomic sampling improves resolution of the animal tree of life. Nature 2008; 452:745 - 749
- Nickel M. DeSalle R, Schierwater B. The Pre-Nervous System and Beyond. Key Transitions in Animal Evolution 2009; Oxford Oxford University Press (in prep.)
- Nickel M. Movements without muscles, information processing without nerves. JMBA GME 2007; 6:8 - 9
- Sakarya O, Armstrong KA, Adamska M, Adamski M, Wang IF, Tidor B, et al. A post-synaptic scaffold at the origin of the animal kingdom. PLoS ONE 2007; 2:506
- Ellwanger K, Nickel M. Neuroactive substances specifically modulate rhythmic body contractions in the nerveless metazoon Tethya wilhelma (Demospongiae, Porifera). Front Zool 2006; 27:3 - 7
- Schierwater B, de Jong D, DeSalle R. Placozoa, and the evolution of Metazoa and intrasomatic cell differentiation. Int J Biochem Cell Biol 2009; 41:370 - 379
- Hadrys T, DeSalle R, Sagasser S, Fischer N, Schierwater B. The trichoplax PaxB gene: a putative proto-PaxA/B/C gene predating the origin of nerve and sensory cells. Mol Biol Evol 2005; 22:1569 - 1578