Abstract
Autophagy is a membrane trafficking pathway responsible for the breakdown of unwanted intracellular materials and crucial for the cell healthiness and survival. In the autophagic flux, various dynamic membrane rearrangements occurs starting with the elongation of the phagophore and its closure to build an autophagosome and ending with its fusion with late endosomes and lysosomes to form an autolysosome. Although Ca2+ is a well established regulator of membrane fusion events, little is known about its role in these processes during autophagy. Recent studies, based on proteomic analyses of lysosomal membranes, have provided new insights into this field of study. Thus, the levels on lysosomal membranes of annexin A1, annexin A5 and copine 1, three proteins that bind to phospholipid membranes in a Ca2+-dependent manner, increased under nutrient deprivation, a condition that promotes autophagic degradation. In addition, two different studies showed that annexin A5 and annexin A1 are involved in autophagosome maturation. Here, we discuss the molecular mechanisms by which the fusion of autophagosomes with endosomes and lysosomes could be regulated by these three proteins and Ca2+.
Background
The clearance of cell components, in particular those that are damaged, is essential for cell welfare and survival. This is mainly, but not exclusively, performed by two different degradation pathways involving proteasomes or lysosomes.Citation1 The delivery of intracellular material to lysosomes for breakdown is principally mediated by double membrane vesicles called autophagosomes. In the last years, the origin of the autophagosomal membrane and the molecular mechanisms of autophagosome formation have been extensively analyzed and discussed. However, much less attention has been paid to later steps in the autophagic process, when autophagosomes deliver their content to acidic compartments for degradation, which are nonetheless the final destiny of the sequestered material. At this stage, autophagosomes mature by fusing with different endocytic and lysosomal vesicles, which add complexity to these fusion events.
Autophagosomal Fusion Machinery
Although the available information on this machinery is still fragmentary, several cell components have been described to be involved in the fusion of autophagosomes with endo-lysosomal compartments. For example, microtubules are thought to direct the traffic of autophagosomes toward lysosomes and endosomes.Citation2,Citation3 Also SNAREs (Soluble N-ethylmaleimide-sensitive factor attachment protein receptors), proteins with a well known role in tethering/docking of vesicles in the presence of Ca2+, Citation4–Citation6 induce, in association with the Rab7 GTPase and the HOPS (Homotypic fusion and protein sorting) complex, the fusion of autophagosomes with lysosomes.Citation7-Citation10 In addition, it has been reported that the three ESCRT (Endosomal sorting complex required for transport) complexes, I–III, which were originally associated with the sorting of ubiquitinated membrane proteins into multivesicular bodies,Citation11-Citation13 participate in the fusion of autophagosomes with lysosomes by mechanisms that are still unknown.Citation14
Involvement of Annexins in Autophagy
Annexins are a family of ubiquitous and Ca2+-dependent membrane-binding proteins whose functions depend on their ability to attach to specific lipid microdomains. Using a proteomic approach, we recently identified annexin A5 as a regulator of autophagosome maturation, especially in the starvation response, where it localizes on lysosomal membranes in a Ca2+-dependent way.Citation15 Under starvation conditions, annexin A5 translocates from the Golgi complex to lysosomes and, to a lesser extent, to late endosomes. Interestingly, this protein was found to inhibit fluid phase and cholera toxin endocytosis. Since annexin A5 localizes in late endosomes considerably more than in early endosomes, it is likely that the observed inhibition of endocytosis occurs at the late steps of this process. Moreover, annexin A5 induces autophagosome fusion with lysosomes, but inhibits the formation of amphisomes, hybrid organelles produced by the fusion of autophagosomes with late endosomes. Although the molecular basis of these two opposite roles of annexin A5, activation of autophagy and inhibition of endocytosis, remains to be elucidated, it is possible that lysosomal and late endosomal membranes have different molecular characteristics in terms of their respective mechanisms of fusion with autophagosomes. In accordance with this concept, at least one protein, Rab7, is required in autophagosome-lysosome fusions,Citation16 but is dispensable in fusions of autophagosomes with late endosomes.Citation17
Experimental findings also support the involvement in autophagy of another protein of the same family, annexin A1. In fact, in the same proteomic analysis we found that, like annexin A5, the levels of annexin A1 increased on lysosomal membranes upon starvation.Citation15 Also, a different group showed that annexin A1 promotes autophagy and suggested that this protein plays a role in the formation of amphisomes.Citation18 Somewhat complementary to these results, a small dimeric Ca2+-binding protein that can form a complex with annexin A1, S100A11, was identified as another component of the lysosomal membrane.Citation19
Mounting evidences support that, in spite of their lack of transmembrane domains, various members of the annexin family can induce membrane fusions.Citation20-Citation22 Thus, several studies (reviewed in Monastyrskaya et al, see ref. 23) described a Ca2+-dependent role of some of these proteins in the formation and traffic of specific endo-lysosomal compartments including annexin A1 (early endosomes, multivesicular bodies and lysosomes) and annexin A5 (late endosomes and lysosomes). Since Ca2+ promotes the fusion of autophagosomes with lysosomes under in vitro conditions,Citation24 these two annexins may be involved in Ca2+-regulated interactions of the autophagosomal and lysosomal membranes that finally lead to their fusion and to the delivery of the autophagosomal content to the lysosome.
Another interesting association of annexin A1 with autophagy was revealed in preliminary maps of interaction networks in autophagy, which are based on a shotgun proteomic analysis.Citation25 These maps identified annexin A1 as a putative interactor of Atg4B. This protease has a crucial role in the formation and maturation of autophagosomes,Citation26,Citation27 because it participates in the conjugation/deconjugation of phosphoethanolamine to LC3, the mammalian ortholog of yeast Atg8. In fact, the balance between lipidation and delipidation of LC3 controls the tethering and hemifusion during closure of the autophagosomal membranes,Citation28 and this is an important requirement for the fusion of autophagosomes with endosomes and lysosomes that occurs later.Citation29 Therefore, it is tempting to speculate that annexin A1 stimulates the fusogenic potential of autophagosomes by regulating the activity of Atg4B.
Possible Involvement in Autophagy of Copine 1 in Relationship with Annexins
Copine 1 is another protein whose levels increased on lysosomal membranes under high proteolysis conditions.Citation15 It shares with the annexin family of proteins the property of binding to phospholipid membranes in a Ca2+-dependent manner.Citation30 In addition, its presence in autophagosomal, phagosomal and lysosomal delimiting membranes has been previously reported.Citation31,Citation32 Interestingly, an in vitro study showed that annexin A1 creates membrane domains enriched in phosphatidyl serine (PS) that assemble copine 1 aggregates. This provides a possible scaffold to cluster signaling proteins in the presence of Ca2+.Citation33
Also, annexin A5 is known to bind with high specificity to PS, using Ca2+ as a bridge between the negatively charged convex side of the C-terminal domain of the protein and the anionic phospholipid.Citation34 PS is known to be distributed in all cellular membranes, but it only confers a negative charge (ideal for Ca2+ binding) at the cytosolic face of endosomes and lysosomes. This is probably because in other organelles, such as mitochondria, Golgi and endoplasmic reticulum, PS is localized in the luminal leaflets of their membranes.Citation35 Although annexins A1 and annexin A5 lack a coiled-coil domain, which according to a yeast two-hybrid screening study facilitates copine 1 binding,Citation36 it is possible that annexin A5, like annexin A1, also forms suitable domains for copine 1 recruitment, after its translocation under high proteolysis conditions from cytosol to lysosomal membranes, to facilitate the fusion of lysosomes with autophagosomes,
Conclusions and Future Work
Annexin A1, annexin A5, and probably also copine 1, emerge as possible regulators of autophagosome maturation by mechanisms that require Ca2+. Both annexins could interact on the lysosomal surface with copine 1, by mechanisms regulated by phospholipid rearrangements and Ca2+ (see ), to promote the fusion of autophagosomes with lysosomes.
Figure 1. Possible mechanisms for the involvement of annexin A5, annexin A1 and copine 1 in autophagosome maturation: The sequence of the different steps in autophagy is shown below. Starvation induces annexin A5 translocation to lysosomal membranes in a Ca2+-dependent way. This protein inhibits (indicated by the blunted red lines) endocytosis and amphisome formation and induces (red arrow) autophagosome fusion with lysosomes. Likewise, two other Ca2+-dependent phospholipid binding proteins, annexin A1 and copine 1, are localized on lysosomal membranes under starvation. It is likely that annexin A5 and annexin A1 aggregate on lysosomal membranes in a Ca2+-dependent way in order to form domains for the subsequent binding of copine 1, and that interactions between these and perhaps other proteins promote the fusion of autophagosomes with lysosomes. An additional possibility, at least for annexin A1, is that its interaction with Atg4B regulates the activity of this protease in the lipidation/delipidation of LC3 and thus contributes to increase the ability of autophagosomal membranes to fuse with lysosomes.
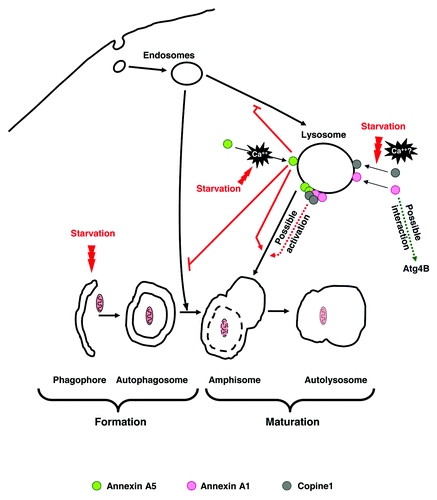
To determine which of the proposed molecular mechanism are involved in this process, it will be necessary to identify other proteins and, perhaps more important, the specific lipids that interact with annexins A1 and annexin A5 and also with copine 1 at the cytosolic surface of the endo-lysosomal membranes. In particular, it would be interesting to find differences in the interacting proteins and lipid domains between lysosomes and late endosomes and also to verify whether these interactions produce phospholipid rearrangements at specific lipid domains.
Acknowledgments
Work in the authors' lab is supported by the Spanish Ministerio de Economía y Competitividad (BFU2008-00186 and BFU2011-22630), Fundació Marató TV3 (ref. 100130) and Centro de Investigación Biomédica en Red de Enfermedades Raras (CIBERER).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Knecht E, Aguado C, Cárcel J, Esteban I, Esteve JM, Ghislat G, et al. Intracellular protein degradation in mammalian cells: recent developments. Cell Mol Life Sci 2009; 66:2427 - 43; http://dx.doi.org/10.1007/s00018-009-0030-6; PMID: 19399586
- Ravikumar B, Acevedo-Arozena A, Imarisio S, Berger Z, Vacher C, O’Kane CJ, et al. Dynein mutations impair autophagic clearance of aggregate-prone proteins. Nat Genet 2005; 37:771 - 6; http://dx.doi.org/10.1038/ng1591; PMID: 15980862
- Fass E, Shvets E, Degani I, Hirschberg K, Elazar Z. Microtubules support production of starvation-induced autophagosomes but not their targeting and fusion with lysosomes. J Biol Chem 2006; 281:36303 - 16; http://dx.doi.org/10.1074/jbc.M607031200; PMID: 16963441
- Parlati F, McNew JA, Fukuda R, Miller R, Söllner TH, Rothman JE. Topological restriction of SNARE-dependent membrane fusion. Nature 2000; 407:194 - 8; http://dx.doi.org/10.1038/35025076; PMID: 11001058
- Atlashkin V, Kreykenbohm V, Eskelinen EL, Wenzel D, Fayyazi A, Fischer von Mollard G. Deletion of the SNARE vti1b in mice results in the loss of a single SNARE partner, syntaxin 8. Mol Cell Biol 2003; 23:5198 - 207; http://dx.doi.org/10.1128/MCB.23.15.5198-5207.2003; PMID: 12861006
- Jena BP. Role of SNAREs in membrane fusion. Adv Exp Med Biol 2011; 713:13 - 32; http://dx.doi.org/10.1007/978-94-007-0763-4_3; PMID: 21432012
- Zerial M, McBride H. Rab proteins as membrane organizers. Nat Rev Mol Cell Biol 2001; 2:107 - 17; http://dx.doi.org/10.1038/35052055; PMID: 11252952
- Sun Q, Westphal W, Wong KN, Tan I, Zhong Q. Rubicon controls endosome maturation as a Rab7 effector. Proc Natl Acad Sci U S A 2010; 107:19338 - 43; http://dx.doi.org/10.1073/pnas.1010554107; PMID: 20974968
- Sato TK, Rehling P, Peterson MR, Emr SD. Class C Vps protein complex regulates vacuolar SNARE pairing and is required for vesicle docking/fusion. Mol Cell 2000; 6:661 - 71; http://dx.doi.org/10.1016/S1097-2765(00)00064-2; PMID: 11030345
- Seals DF, Eitzen G, Margolis N, Wickner WT, Price AA. A Ypt/Rab effector complex containing the Sec1 homolog Vps33p is required for homotypic vacuole fusion. Proc Natl Acad Sci U S A 2000; 97:9402 - 7; http://dx.doi.org/10.1073/pnas.97.17.9402; PMID: 10944212
- Katzmann DJ, Babst M, Emr SD. Ubiquitin-dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex, ESCRT-I. Cell 2001; 106:145 - 55; http://dx.doi.org/10.1016/S0092-8674(01)00434-2; PMID: 11511343
- Babst M, Katzmann DJ, Snyder WB, Wendland B, Emr SD. Endosome-associated complex, ESCRT-II, recruits transport machinery for protein sorting at the multivesicular body. Dev Cell 2002; 3:283 - 9; http://dx.doi.org/10.1016/S1534-5807(02)00219-8; PMID: 12194858
- Babst M, Katzmann DJ, Estepa-Sabal EJ, Meerloo T, Emr SD. Escrt-III: an endosome-associated heterooligomeric protein complex required for mvb sorting. Dev Cell 2002; 3:271 - 82; http://dx.doi.org/10.1016/S1534-5807(02)00220-4; PMID: 12194857
- Rusten TE, Stenmark H. How do ESCRT proteins control autophagy?. J Cell Sci 2009; 122:2179 - 83; http://dx.doi.org/10.1242/jcs.050021; PMID: 19535733
- Ghislat G, Aguado C, Knecht E. Annexin A5 stimulates autophagy and inhibits endocytosis. J Cell Sci 2012; 125:92 - 107; http://dx.doi.org/10.1242/jcs.086728; PMID: 22266906
- Ganley IG, Wong PM, Gammoh N, Jiang X. Distinct autophagosomal-lysosomal fusion mechanism revealed by thapsigargin-induced autophagy arrest. Mol Cell 2011; 42:731 - 43; http://dx.doi.org/10.1016/j.molcel.2011.04.024; PMID: 21700220
- Fader CM, Sánchez D, Furlán M, Colombo MI. Induction of autophagy promotes fusion of multivesicular bodies with autophagic vacuoles in k562 cells. Traffic 2008; 9:230 - 50; http://dx.doi.org/10.1111/j.1600-0854.2007.00677.x; PMID: 17999726
- Kang JH, Li M, Chen X, Yin XM. Proteomics analysis of starved cells revealed Annexin A1 as an important regulator of autophagic degradation. Biochem Biophys Res Commun 2011; 407:581 - 6; http://dx.doi.org/10.1016/j.bbrc.2011.03.067; PMID: 21420379
- Nylandsted J, Becker AC, Bunkenborg J, Andersen JS, Dengjel J, Jäättelä M. ErbB2-associated changes in the lysosomal proteome. Proteomics 2011; 11:2830 - 8; http://dx.doi.org/10.1002/pmic.201000734; PMID: 21674799
- McNeil AK, Rescher U, Gerke V, McNeil PL. Requirement for annexin A1 in plasma membrane repair. J Biol Chem 2006; 281:35202 - 7; http://dx.doi.org/10.1074/jbc.M606406200; PMID: 16984915
- Rafikova ER, Melikov K, Ramos C, Dye L, Chernomordik LV. Transmembrane protein-free membranes fuse into xenopus nuclear envelope and promote assembly of functional pores. J Biol Chem 2009; 284:29847 - 59; http://dx.doi.org/10.1074/jbc.M109.044453; PMID: 19696024
- Shen S, Tobery CE, Rose MD. Prm3p is a pheromone-induced peripheral nuclear envelope protein required for yeast nuclear fusion. Mol Biol Cell 2009; 20:2438 - 50; http://dx.doi.org/10.1091/mbc.E08-10-0987; PMID: 19297527
- Monastyrskaya K, Babiychuk EB, Draeger A. The annexins: spatial and temporal coordination of signaling events during cellular stress. Cell Mol Life Sci 2009; 66:2623 - 42; http://dx.doi.org/10.1007/s00018-009-0027-1; PMID: 19381436
- Koga H, Kaushik S, Cuervo AM. Altered lipid content inhibits autophagic vesicular fusion. FASEB J 2010; 24:3052 - 65; http://dx.doi.org/10.1096/fj.09-144519; PMID: 20375270
- Behrends C, Sowa ME, Gygi SP, Harper JW. Network organization of the human autophagy system. Nature 2010; 466:68 - 76; http://dx.doi.org/10.1038/nature09204; PMID: 20562859
- Satoo K, Noda NN, Kumeta H, Fujioka Y, Mizushima N, Ohsumi Y, et al. The structure of Atg4B-LC3 complex reveals the mechanism of LC3 processing and delipidation during autophagy. EMBO J 2009; 28:1341 - 50; http://dx.doi.org/10.1038/emboj.2009.80; PMID: 19322194
- Weidberg H, Shvets E, Shpilka T, Shimron F, Shinder V, Elazar Z. LC3 and GATE-16/GABARAP subfamilies are both essential yet act differently in autophagosome biogenesis. EMBO J 2010; 29:1792 - 802; http://dx.doi.org/10.1038/emboj.2010.74; PMID: 20418806
- Nakatogawa H, Ichimura Y, Ohsumi Y. Atg8, a ubiquitin-like protein required for autophagosome formation, mediates membrane tethering and hemifusion. Cell 2007; 130:165 - 78; http://dx.doi.org/10.1016/j.cell.2007.05.021; PMID: 17632063
- Rusten TE, Vaccari T, Lindmo K, Rodahl LM, Nezis IP, Sem-Jacobsen C, et al. ESCRTs and Fab1 regulate distinct steps of autophagy. Curr Biol 2007; 17:1817 - 25; http://dx.doi.org/10.1016/j.cub.2007.09.032; PMID: 17935992
- Creutz CE, Tomsig JL, Snyder SL, Gautier MC, Skouri F, Beisson J, et al. The copines, a novel class of C2 domain-containing, calcium-dependent, phospholipid-binding proteins conserved from Paramecium to humans. J Biol Chem 1998; 273:1393 - 402; http://dx.doi.org/10.1074/jbc.273.3.1393; PMID: 9430674
- Øverbye A, Fengsrud M, Seglen PO. Proteomic analysis of membrane-associated proteins from rat liver autophagosomes. Autophagy 2007; 3:300 - 22; PMID: 17377489
- Damer CK, Bayeva M, Hahn ES, Rivera J, Socec CI. Copine A, a calcium-dependent membrane-binding protein, transiently localizes to the plasma membrane and intracellular vacuoles in Dictyostelium. BMC Cell Biol 2005; 6:46; http://dx.doi.org/10.1186/1471-2121-6-46; PMID: 16343335
- Creutz CE, Edwardson JM. Organization and synergistic binding of copine I and annexin A1 on supported lipid bilayers observed by atomic force microscopy. Biochim Biophys Acta 2009; 1788:1950 - 61
- Gerke V, Moss SE. Annexins: from structure to function. Physiol Rev 2002; 82:331 - 71; PMID: 11917092
- Yeung T, Gilbert GE, Shi J, Silvius J, Kapus A, Grinstein S. Membrane phosphatidylserine regulates surface charge and protein localization. Science 2008; 319:210 - 3; http://dx.doi.org/10.1126/science.1152066; PMID: 18187657
- Tomsig JL, Snyder SL, Creutz CE. Identification of targets for calcium signaling through the copine family of proteins. Characterization of a coiled-coil copine-binding motif. J Biol Chem 2003; 278:10048 - 54; http://dx.doi.org/10.1074/jbc.M212632200; PMID: 12522145