Abstract
Bach1 is a transcriptional repressor which modulates several critical transcriptional responses, such as the expression of the heme oxygenase-1 (HO-1) gene in response to oxidative stress. In our recent study, we found that Bach1 possesses a novel role in mitotic chromosome alignment during metaphase. Upon BACH1 depletion in HeLa cells, mitotic chromosomes become unstable. This defect was efficiently rescued by expressing Bach1 fragments that lack the DNA binding domain, indicating that its function in mitosis involves a transcription-independent mechanism. The nuclear export signal (NES/CLS) of Bach1 is required for the mitotic function. Bach1 is excluded from the mitotic chromosomes depending on its NES/CLS and the nuclear exporter Crm1. Our findings suggest that Bach1 might mediate the regulation of mitotic chromosomes under conditions of cellular stress.
Regulation of the Subcellular Localization of Bach1
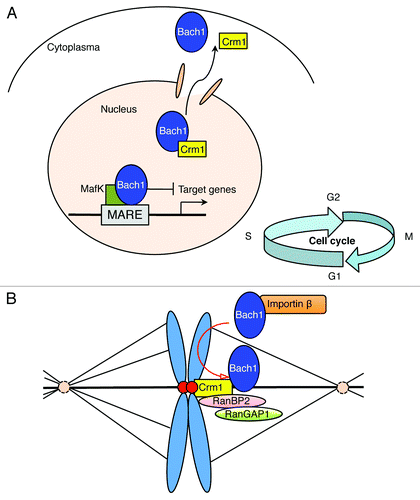
Bach1 (BTB and CNC homology 1) is a transcriptional repressor, the target genes of which include heme oxygenase-1 (HO-1) and globin genes. HO-1 promotes iron recycling by degrading heme into ferrous iron, carbon monoxide (CO) and biliverdin, which is rapidly reduced to bilirubin. Because bilirubin and CO possess cytoprotective roles and heme is a potent prooxidant, HO-1 protects cells from diverse stresses.Citation1,Citation2 Unlike other transcription factors, the subcellular localization of Bach1 is regulated by multiple mechanisms. First, Bach1 possesses an exportin1/Crm1-dependent nuclear export signal (NES) whose activity is activated by direct heme binding ().Citation3 Second, Bach1 possesses another NES at its C-terminus. This signal, originally discovered on Bach1-related factor Bach2 as a cytoplasmic localization signal (CLS), is activated by cadmium in an ERK-dependent manner.Citation4,Citation5 Thus, Bach1 responds to both heme and cadmium, two potent inducers of HO-1, by distinct mechanisms. Third, Bach1 interacts with the cytoplasmic, microtubule-associated, intracellular hyaluronic acid binding protein IHABP, also known as the receptor for hyaluronan-mediated motility (RHAMM).Citation6 IHABP has been suggested to play a role in the organization of the cytoskeletal network by interacting with microtubules and actin filaments, and thus, plays a role in cell morphology and motility.Citation7 It has been unclear why the subcellular localization of Bach1 is regulated by multiple mechanisms. However, it appears that each of the mechanisms may mediate a response to particular signals. Alternatively, some of the mechanisms may connect Bach1 with non-transcriptional functions, which we discuss below.
Figure 1. A model of the dual functions of Bach1 during interphase and metaphase of the cell cycle. (A) Bach1 occupies MARE enhancers to repress transcription under normal conditions. The elimination of the Bach1-mediated transcriptional repression occurs through inhibition of its DNA binding activity and subsequent Crm1-dependent nuclear export in response to oxidative stress. (B) The kinetochore localization of Crm1 mediates the exclusion of Bach1 from mitotic chromosomes and may be required for the kinetochore recruitment of RanBP2 and RanGAP1. We hypothesize that the misalignment of mitotic chromosomes due to Bach1 depletion might involve an improper kinetochore localization of RanGAP1, RanBP2 and/or Crm1.
Bach1 as a Regulator of Mitotic Chromosomes at Metaphase
Arnaoutov et al.Citation8 first reported the behavior of Crm1 during mitosis and showed that Crm1 is a mitotic effector of Ran-GTP in somatic cells. Crm1 and Ran-GTP are essential for the stable recruitment of Ran-GAP1/Ran-BP2 to kinetochores, the formation of normal kinetochore fibers, and faithful chromosome segregation.Citation8 Considering that both of the nuclear export signals of Bach1 are Crm1-dependent, and the IHABP may connect Bach1 to tubulin, we hypothesized that Bach1 might play a role during mitosis through a Crm1-dependent mechanism beyond its canonical transcriptional function.
In our recent study, we discovered a novel role of Bach1 in the regulation of mitotic chromosome dynamics. To examine the role of Bach1 in mitosis, we used short hairpin RNA (shRNA) to deplete endogenous BACH1 in HeLa cells. Surprisingly, the BACH1 depletion resulted in disordered mitotic chromosome alignment, including a thicker mitotic chromosome plate, collapsed ring-like shape and oscillating chromosome axis. These defects were efficiently rescued by not only wild-type mouse Bach1, but also its derivatives lacking the DNA binding domain, suggesting that the function of Bach1 in mitosis involves a transcription-independent mechanism.Citation9 Furthermore, Bach1 derivatives lacking the C-terminal NES/CLS failed to rescue the mitotic defects in BACH1-depleted HeLa cells, thus suggesting the involvement of Crm1 in this process. Consistent with this prediction, Bach1 was excluded from mitotic chromosomes in a manner depending on both CLS and Crm1.
Unexpectedly, the misalignment of chromosomes due to the depletion of BACH1 did not cause a fatal cell cycle failure. This result suggests that the abnormal mitotic chromosomes observed in the BACH1-depleted cells reflect a transient process which can be corrected immediately by a redundant, compensating activity. The time-lapse analysis of BACH1-depleted cells revealed dynamic changes of the mitotic chromosomes. BACH1-depleted cells showed the transient formation of a thicker mitotic chromosome plate and misorientation of the mitotic axis. Moreover, these thicker mitotic chromosomes and their axes oscillated in the proximity of the lateral cell cortex.Citation9
The Mitotic Role of Bach1 Might Involve a Dynein-dependent Mechanism
These observations strongly suggest that Bach1 is involved in the regulation of mitotic chromosome formation and its dynamics. Although the molecular basis for this regulation is still unclear, the findings of previous reports suggest several interesting possibilities. First, the cortical dynein localization is regulated by two distinct intrinsic signals to determine the spindle position and orientation in symmetrically dividing human cells: a spindle pole-derived signal of Plk1 and a chromosome-derived gradient of RanGTP.Citation10 On the other hand, inhibition of phosphatidylinositol-3-OH kinase (PI(3)K) causes dynein-dependent spindle rotations along the z-axis, resulting in spindle misorientation.Citation11 Taken together, these observations suggest that a dynein-dynactin-dependent pulling force on astral microtubules is essential for maintaining the spindle orientation. Surprisingly, we found that BACH1 interacts with DYNLL1 (dynein light chain 1) according to the MitoCheck database (www.mitocheck.org). Therefore, future studies should examine how Bach1 interacts with dynein to regulate the mitotic chromosome alignment.
Possible Involvement of Bach1 in the RANBP2-CRM1 Pathway
Importin-β is the main vector for protein import in interphase nuclei.Citation12 It also has roles after nuclear envelope breakdown through binding with RANGTP to release some NLS-containing mitotic factors, and thus functions in spindle organization.Citation13 A recent study showed that overexpression of importin-β induces mitotic dynamic defects, including multi-polar division and unstable chromosome alignment called “a broader plate.” This defect appears to be due to the inhibition of RANGAP1 recruitment to mitotic kinetochores, an event that is known to require microtubule attachment and the exportin, CRM1.Citation8,Citation14 Co-expression with RANBP2 or CRM1 suppresses the importin-β-induced mitotic dynamic defects. Interestingly, this “broader plate” phenotype is similar to that of BACH1-depleted HeLa cells. Therefore, the mitotic chromosome misalignment due to BACH1 depletion might involve an improper kinetochore localization of RANGAP1 and/or CRM1 ().
Proteomic Analysis of the Protein Network of Bach1
To clarify the molecule(s) that link Bach1 in mouse cells to the regulation of mitotic chromosomes, we are currently investigating the protein network of Bach1 by a proteomic analysis. Among the various proteins found to interact with Bach1, ATM (unpublished data) is one of the critical regulators of the DNA damage response (DDR). Another interesting protein is Bub1 (unpublished data) which is a key kinase involved in spindle checkpoint activation. In addition to the classical DDR function, ATM is also activated by Aurora-B during mitosis. The mitotic activation of ATM is required for the activation of Bub1, thus regulating the spindle checkpoint. It should be noted that this novel role of ATM is separable from the DDR.Citation15 Taken together, these findings warrant further studies about the molecular mechanism underlying how Bach1 regulates chromosome alignment during mitosis.
Mitosis is the shortest but most pivotal phase of the cell cycle, and gives rise to identical or distinct pairs of cells. Although it has a well-known role as a transcriptional regulator, our recent studies have, for the first time, revealed a novel function of Bach1 for the regulation of mitotic chromosome alignment and the proper orientation of the mitotic axis that is independent of its transcriptional effects. Clarification of this mechanism involving Bach1 might be helpful in understanding various processes, such as stem cell differentiation and cancer progression.
| Abbreviations: | ||
| Bach1 | = | BTB and CNC homology 1 |
| Crm1 | = | chromosome region maintenance protein 1 |
| HO-1 | = | heme oxygenase-1 |
| NES | = | nuclear export signal |
| CLS | = | cytoplasmic localization signal |
Acknowledgments
We would like to thank Mr. Hiroki Shima for various suggestions. This study was supported by Grants-in-Aid and the Network Medicine Global COE Program from the Ministry of Education, Culture, Sport, Science and Technology of Japan. The authors declare that they have no conflict of interest.
References
- Igarashi K, Sun J. The heme-Bach1 pathway in the regulation of oxidative stress response and erythroid differentiation. Antioxid Redox Signal 2006; 8:107 - 18; http://dx.doi.org/10.1089/ars.2006.8.107; PMID: 16487043
- Sun J, Hoshino H, Takaku K, Nakajima O, Muto A, Suzuki H, et al. Hemoprotein Bach1 regulates enhancer availability of heme oxygenase-1 gene. EMBO J 2002; 21:5216 - 24; http://dx.doi.org/10.1093/emboj/cdf516; PMID: 12356737
- Suzuki H, Tashiro S, Hira S, Sun J, Yamazaki C, Zenke Y, et al. Heme regulates gene expression by triggering Crm1-dependent nuclear export of Bach1. EMBO J 2004; 23:2544 - 53; http://dx.doi.org/10.1038/sj.emboj.7600248; PMID: 15175654
- Suzuki H, Tashiro S, Sun J, Doi H, Satomi S, Igarashi K. Cadmium induces nuclear export of Bach1, a transcriptional repressor of heme oxygenase-1 gene. J Biol Chem 2003; 278:49246 - 53; http://dx.doi.org/10.1074/jbc.M306764200; PMID: 14504288
- Hoshino H, Kobayashi A, Yoshida M, Kudo N, Oyake T, Motohashi H, et al. Oxidative stress abolishes leptomycin B-sensitive nuclear export of transcription repressor Bach2 that counteracts activation of Maf recognition element. J Biol Chem 2000; 275:15370 - 6; http://dx.doi.org/10.1074/jbc.275.20.15370; PMID: 10809773
- Yamasaki C, Tashiro S, Nishito Y, Sueda T, Igarashi K. Dynamic cytoplasmic anchoring of the transcription factor Bach1 by intracellular hyaluronic acid binding protein IHABP. J Biochem 2005; 137:287 - 96; http://dx.doi.org/10.1093/jb/mvi031; PMID: 15809329
- Gundersen GG, Cook TA. Microtubules and signal transduction. Curr Opin Cell Biol 1999; 11:81 - 94; http://dx.doi.org/10.1016/S0955-0674(99)80010-6; PMID: 10047525
- Arnaoutov A, Azuma Y, Ribbeck K, Joseph J, Boyarchuk Y, Karpova T, et al. Crm1 is a mitotic effector of Ran-GTP in somatic cells. Nat Cell Biol 2005; 7:626 - 32; http://dx.doi.org/10.1038/ncb1263; PMID: 15908946
- Li J, Shiraki T, Igarashi K. Transcription-independent role of Bach1 in mitosis through a nuclear exporter Crm1-dependent mechanism. FEBS Lett 2012; 586:448 - 54; http://dx.doi.org/10.1016/j.febslet.2012.01.028; PMID: 22289179
- Kiyomitsu T, Cheeseman IM. Chromosome- and spindle-pole-derived signals generate an intrinsic code for spindle position and orientation. Nat Cell Biol 2012; 14:311 - 7; http://dx.doi.org/10.1038/ncb2440; PMID: 22327364
- Toyoshima F, Matsumura S, Morimoto H, Mitsushima M, Nishida E. PtdIns(3,4,5)P3 regulates spindle orientation in adherent cells. Dev Cell 2007; 13:796 - 811; http://dx.doi.org/10.1016/j.devcel.2007.10.014; PMID: 18061563
- Harel A, Forbes DJ. Importin beta: conducting a much larger cellular symphony. Mol Cell 2004; 16:319 - 30; http://dx.doi.org/10.1016/S1097-2765(04)00647-1; PMID: 15525506
- Nachury MV, Maresca TJ, Salmon WC, Waterman-Storer CM, Heald R, Weis K. Importin beta is a mitotic target of the small GTPase Ran in spindle assembly. Cell 2001; 104:95 - 106; http://dx.doi.org/10.1016/S0092-8674(01)00194-5; PMID: 11163243
- Roscioli E, Di Francesco L, Bolognesi A, Giubettini M, Orlando S, Harel A, et al. Importin-β negatively regulates multiple aspects of mitosis including RANGAP1 recruitment to kinetochores. J Cell Biol 2012; 196:435 - 50; http://dx.doi.org/10.1083/jcb.201109104; PMID: 22331847
- Yang CY, Tang X, Guo XJ, Niikura Y, Kitagawa K, Cui KM, et al. Aurora-B mediated ATM serine 1403 phosphorylation is required for mitotic ATM activation and the spindle checkpoint. Mol Cell 2011; 44:597 - 608; http://dx.doi.org/10.1016/j.molcel.2011.09.016; PMID: 22099307