Abstract
Two of the most widely and intensively cultivated jute species, Corchorus capsularis and Corchorus olitorius, suffer severely from a stem rot disease caused by the fungus Macrophomina phaseolina. Wild jute species, C. trilocularis, shows resistance to this pathogenic fungus. In this study, the technique of differential display was applied to identify genes which are differentially expressed, under both infected and un-infected conditions, between C. trilocularis and C. olitorius var O-72. Two xyloglucan endotransglycosylase/hydrolase (XTH) genes designated CoXTH1 (from Corchorus olitorius) and CtXTH1 (from C.trilocularis) were identified from each of the two species which show different expression patterns upon fungal infection. A steady rise in the expression of CtXTH1 in response to infection was observed by quantitative real time PCR whereas the expression of CoXTH1 was found to be downregulated. Full length sequences of these two genes were determined using primer based gene walking and RACE PCR. This study confirms the involvement of XTH in molecular interactions between M. phaseolina and jute. However, it remains to be explored whether XTH is an essential component of the signaling pathway involved in plant-fungal interaction.
Keywords: :
Introduction
Both Corchorus capsularis and Corchorus olitorius suffer severely from stem rot and charcoal rot, caused by a pathogen Macrophomina phaseolina.Citation1 Other than jute this phytopathogenic fungus infects more than 500 cultivated and wild plant species and has a wide geographic distribution.Citation2,Citation3 The pathogen can cause infection and damage at all stages of plant growth, from seedling emergence right up to maturation. M. phaseolina survives as microsclerotia in the soil and on infected plant debris that serves as the primary source of inoculum.Citation4 It disrupts nutrient and water transport to the upper parts of the plant as it affects the vascular system of the roots and basal internodes leading to wilting, premature death and reduced yield. It also causes root rot, basal stem rot and seedling blight.Citation4,Citation5 Since the yield of jute is severely reduced, development of a variety that shows resistance to M. phaseolina will be of great importance to the countries growing this economically important fiber crop.
Wild jute species, C. trilocularis shows resistance to M. phaseolina,Citation6 while C. olitorius var O-72, a widely cultivated variety, is susceptible to it. We have studied differential gene expression between these two species under both infected and uninfected conditions at different time intervals using a technique called mRNA differential displayCitation7 and have identified two differentially regulated xyloglucan endotransglycosylase/hydrolase genes (CtXTH1 in C. trilocularis and CoXTH1 in C. olitorius) which show difference in their expression patterns upon fungal infection.
Because of their inherent resistance capacity plants have overcome numerous challenges from different pathogens. Before encountering the intracellular defense, a pathogen has to face the cell wall, which has as an important role in plant defense. Upon attack, various defense related polymers are synthesized to reinforce the cell wall at the sites, where the pathogens attempt penetration.Citation8 XTHs are one of the major players in this process as they cause rearrangement of the cellulose/xyloglucan architecture in the cell wall. The enzyme catalyzes transfer of a segment of one xyloglucan molecule allowing for molecular grafting between the polysaccharide molecules that subsequently change both the cell wall plasticity and architecture.Citation9-Citation11 There is evidence that one of the key interactions in the primary wall of dicotyledons takes place between cellulose and the hemicellulose xyloglucan, which together typically comprise about two thirds of the dry wall mass. Xyloglucan binds non-covalently to cellulose, coats and cross-links adjacent cellulose microfibrils and the resulting extensive xyloglucan-cellulose network is thought to act as the principal tension-bearing structure in the cell wall.Citation12-Citation14 Our current understanding suggests that, either the breakage of xyloglucan cross-links or their detachment from cellulose microfibrils is necessary for microfibrils to move apart to ensure cell wall expansion.Citation15,Citation16 Several homologs of XTH have been identified in different plant species. They constitute a large multi-gene family designated as xyloglucan-related proteins (XRP).Citation16,Citation17 Although the key function of XTH is cell wall modification, the existence of many different isozymes and their different temporal and spatial expression indicates additional roles other than cell wall growth and elongation.
Results
Identification of differentially expressed XTH gene and its phylogenetic analyses
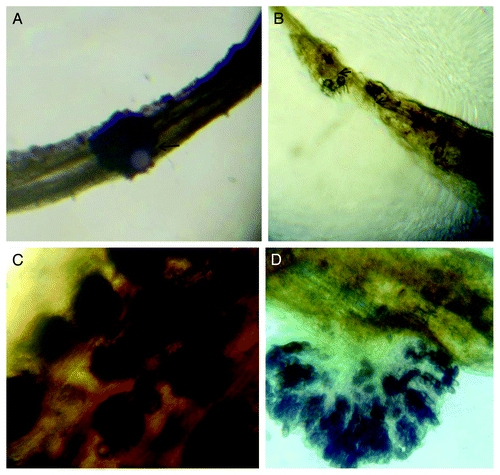
In order to study the interaction between jute (Corchorus spp) and M. phaseolina at the molecular level, we inoculated jute seedlings with M. phaseolina sclerotia. C. olitorius, the susceptible species, started showing diseased symptoms such as browning of the root within 24 h of infection. To confirm the sclerotia infection, we examined the root tissue sections under a light microscope. Sclerotial attachment was observed in the samples collected 15 and 24 h after inoculation, in both the species (figure not shown). However, two days after incubation, the fungus showed signs of growth and colonization in C. trilocularis roots (). Germination of the hyphae together with its penetration (), formation of sclerotia in root tissues () and conidia formation and local destruction of host tissue () could be seen under the microscope. The inoculation process we followed in the laboratory is perhaps different from the actual infection process in the field; nevertheless this method can be an effective pathosystem to study molecular interactions between jute and Macrophomina.
Figure 1.M. phaseolina infected seedlings after two days of infection. (A) C. trilocularis seedlings, sclerotia attached to the root (B) Both intra and inter cellular Hyphal growth in root and subsequently growth of sclerotia in C. olitorius seedlings (C) New sclerotia (dark spots) observed in infected C. olitorius seedlings (D) Conidia formation and destruction of the host tissue (in C. olitorius)
The transcript profiles of infected and uninfected C. olitorious (var: O-72) and C. trilocularis seedlings were compared by mRNA differential display. Total RNA from these plants were collected and their quality and quantity were determined. Quality of isolated RNA was checked by electrophoresis (data not shown). In order to check the reproducibility of the method used, two separate mRNA differential display RT-PCR were performed and, almost equal number of bands was produced (Table S1) both the times. Some of the differentially expressed genes were amplified in all the three samples (S0, S15 and S24) from susceptible plants but none from the resistant ones. The opposite was also observed. In other cases, amplifications were found only in samples after a certain period of infection. Therefore, both constitutive and induced patterns of differential gene expression can be studied following the technique of differential display. In our study, we focused on one particular differentially expressed gene (starting with a cDNA fragment of 246bp from C. trilocularis). The amplified product of this gene was recovered from the gel, sub-cloned and sequenced. BLASTn and BLASTx analyses indicated the sequence to be a putative XTH gene.
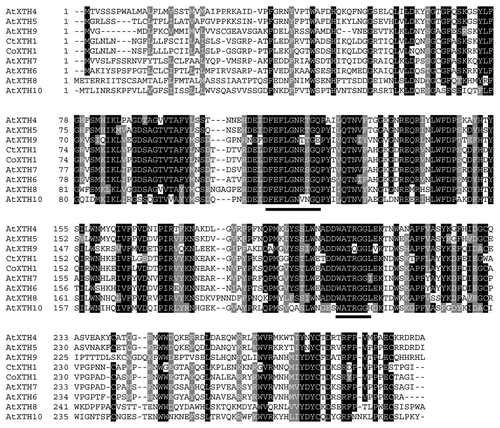
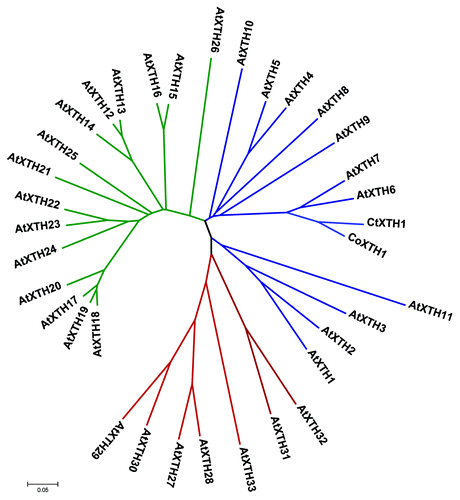
The full length sequences of CtXTH1 and CoXTH1 were determined using primer-based gene walking and conventional 3′ and 5′ RACE. PCR of genomic DNA helped to identify the introns. Both the genes were found to have three introns and the coding region was 873 bp long (), having a similarity of 92% (BLASTn between two sequences). SignalPCitation18 analysis shows that for both the proteins, first 26 amino acids act as signal peptide to guide the same to the cell wall. Rose et al.,Citation14 classified Arabidopsis XTHs into three major groups. Phylogenetic analysis using 33 Arabidopsis XTHs have classified CtXTH1 and CoXTH1 with group I genes. As shown in , they are placed in the same clade with seven of the Arabidopsis group I genes (AtXTH 10, AtXTH 5, AtXTH4, AtXTH 8, AtXTH 9, AtXTH 7 and AtXTH 6). Michailidis and coworkers Citation19 classified the rest of the group I genes (AtXTH 1, AtXTH 2, AtXTH 3 and AtXTH 11) as ancestral group and they are grouped in a different clade.
Figure 2. Schematic diagram of CtXTH1 and CoXTH1 gene structure, the white boxes represent exons, lines introns and the shaded boxes are the putative signal peptide sequences.
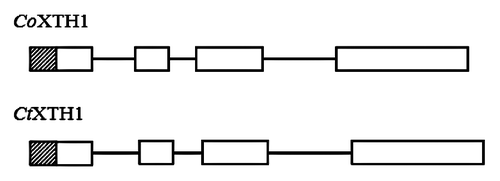
Figure 3. Phylogenetic relationship of CoXTH1 and CtXTH1 with Arabidopsis XTH genes. The tree was generated by MEGA 4.1 using neighbor-joining method and p-distance correction. The Arabidopsis genes from Group 1, 2 and 3 are shown in blue, green and red, respectively.
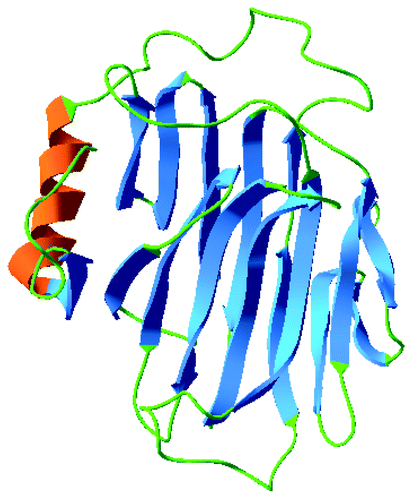
The conserved motif, DEIDFELFG, described as the catalytic siteCitation17 appears in CtXTH1 and in CoXTH1 as DELDFELFG, whereas, the sequence of the motif for the seven Arabidopsis group I genes is found to be DE(I/F/L)DFELFG in the multiple sequence alignment (). In addition, the motif DWATRGG that ensures the hydrophobic stacking interactions with glucose unit is also present.Citation20 The 3D model of CtXTH1 () constructed using CPHmodels-3.0 server and represented using Swiss-Pdb ViewerCitation21 shows β-jellyroll-type structure typical of other GH16 family enzymes.Citation22
CtXTH1 and CoXTH1 are members of a large gene family
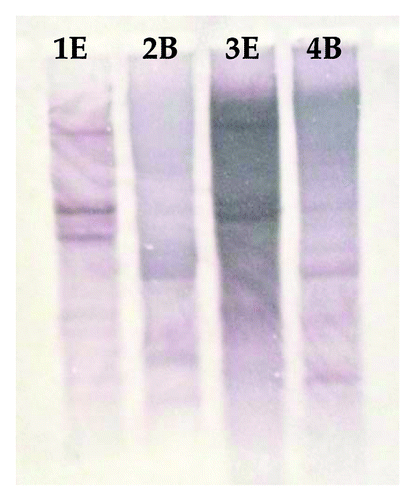
To determine the copy number of XTHs in C. olitorious and C. trilocularis, gDNA from both the plant species was digested with EcoRI and BamHI as these restriction enzymes do not have a recognition site within CtXTH1 and CoXTH1 genes. Digested gDNA was hybridized with cDNA derived probes at low stringency. For both CtXTH1 and CoXTH1, more than one band was -detected implying that they are members of a large gene family ().
CtXTH1 and CoXTH1 shows different expression profile upon M. phaseolina infection
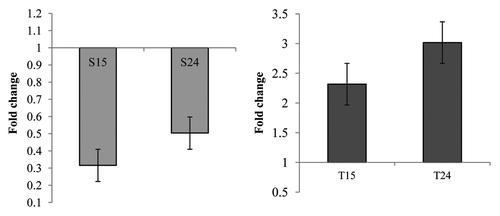
To estimate the expression patterns of these two genes, expression analysis was performed using semi-quantitative RT-PCR and real-time qPCR with a pair of primers which amplify the same region in both genes. Initially, we expected the expression of XTH in the sensitive plant (CoXTH1 in C. olitorius) would be higher after fungal infection and the XTH from the tolerant plant (CtXTH1 in C. trilocularis) should show a different pattern. To our surprise, the results indicated that CtXTH1 expression is upregulated following fungal infection whereas CoXTH1 shows downregulation ( and ). CoXTH1 expression was found to decrease at the initial time point (15 h) compared with the uninfected sample (S0 expression level) followed by a slight increase at 24 hrs. However, the initial expression level of CtXTH1 (untreated C. trilocularis seedlings, T0) was much higher compared with CoXTH1 and the highest level of CtXTH1 expression was observed after 24 h of infection ().
Figure 7. (A) Semi-quantitative Reverse Transcription-PCR expression analysis of three C. olitorious samples (S0, S15 and S24) and three C. trilocularis samples (T0, T15 and T24) using XTHG'F and XTH 3′R primers. (B) The housekeeping gene β-actin was used to verify the quantitation.
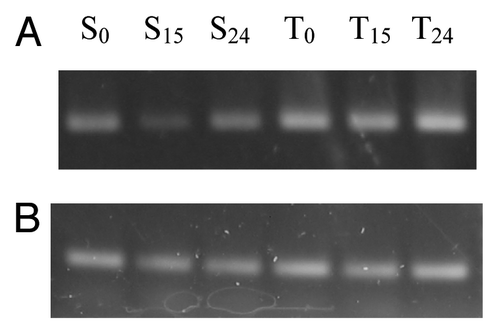
Figure 8. Real-time PCR analysis of (A) CoXTH1 and (B) CtXTH1 genes to see the change in expression level by M. phaseolina. Samples were infected and collected after 15 (S15 and T15) and 24 (S24 and T24) hours of incubation. The mean fold changes of expression (of two duplicate assays) using RNA samples from infected seedlings shows the downregulation of CoXTH1 and upregulation of CtXTH1 followed by fungal infection.
Discussion
Till date, little is known about the molecular mechanisms behind the pathogenicity of M. phaseolina. One way to comprehend these molecular interactions is to look at the gene expression pattern(s). It is possible that there is a certain correlation between the pathogenicity of the fungus, its ability to invade the host and the different gene expression profiles of the tolerant and sensitive plant species. Both constitutive and induced gene expression, especially at the early stages of fungal infection, could therefore explain the response of the host to the invading pathogen. It could elucidate, albeit partially, the level of tolerance or sensitivity against the pathogen and the physiological responses displayed by the host. In line with this reasoning, this study aimed to look at the gene expression pattern of a tolerant and sensitive species using a technique called differential display.Citation23
We have identified two orthologs, one from C. olitorius and another from C. trilocularis which show differential expression upon fungal infection. Both the genes share a high homology with XTH genes of Arabidopsis group 1 subfamily () and the intron-exon patterns are typical of previously annotated XTH genes. To the best of our knowledge, CtXTH1 is the only gene from C. trilocularis that has been completely sequenced and characterized (GenBank accession: GU809232)
Due to its association with many physiological and developmental processes, XTH gene family is perhaps one of the few plant gene families that have been extensively studied. In every species, this family has expanded and through evolutionary changes, each member of the gene family within a species has attained the capability of different spatial and temporal expression. Prior studies have shown that duplication has been the primary mediator of XTH family expansion in ArabidopsisCitation24 and it will not be surprising, if like other plants the same is also found true for jute. Although there are numerous studies on XTH genes, relationship between their expression pattern and function is still a paradox. Previous studies have shown that 33 Arabidopsis XTH genes exhibit different organ specific expression pattern and respond differently to different sets of plant hormones.Citation24-Citation26 Generally, XTHs are thought to play two distinct rolesCitation27,Citation28 which are necessary for cell wall expansion. One is ‘integrational’ transglycosylation and the other is ‘restructuring’ transglycosylation. The ‘integrational’ transglycosylation is a wall strengthening function of XTH where a newly synthesized and secreted xyloglucan chain is grafted to existing wall bound xyloglucan.Citation29 On the other hand, ‘restructuring’ type of transglycosylation leads to cutting and joining of xyloglucan chains bound previously to the cell wall.Citation25 This activity is of particular importance as it causes wall-strengthening. Takeda and coworkersCitation30 have shown that addition of xyloglucan to the excised pea stems causes it to become stiffer.Citation30
Previous physiological studies have indicated that compared with its high yielding counterpart C. olitorious, C. trilocularis is relatively sturdy and more adaptive.Citation6 It is possible that the high XTH expression in C. trilocularis is one of the major players in causing the cell wall stiffness that eventually acts against fungal invasion. Additionally, regulation of XTH expression can be modulated by hormonal response and the physical organization of the genes in a genome.Citation31,Citation32 Earlier studies have also shown that XTH genes demonstrate a broad range of responses to environmental stresses, both biotic and abiotic.Citation17,Citation33-Citation35 XTH expression is also correlated with cell elongation, as observed in Arabidopsis shootsCitation17,Citation36 and root tips.Citation37 Albert and coworkersCitation25 found the tomato XTH (LeXTH1) expression level to increase upon parasitic attack however mechanical wounding was found to have no effect. Another group, Maldonado-Mendoza et al.,Citation31 found XTH activity to be induced systemically in mycorrhizal roots. In our case, even though both the enzymes were found to show high homology at the sequence level, but the difference in their expression pattern was possibly due to a distinct pattern of their regulation. This could be the outcome of (i) a difference in the accumulation and fixation of mutations between the two species, or (ii) a hormonal difference in the control of the regulon of CoXTH1 and CtXTH1. However, we did not find any positive Darwinian selection between the two genes when we analyzed the substitution pattern. The codon based Z test also indicated neutrality for most (91.29%) of the pair wise comparisons of Arabidopsis XTH genes (data not shown). So, the different regulatory pattern of XTH genes may have arisen from gene duplication and evolutionary diversification that ensured their tissue and hormone specific (spatial and temporal) expression patterns. It is evident from the current study that CoXTH1 and CtXTH1 genes in C. olitorius and C. trilocularis are members of large gene families () and it is possible that due to evolutionary diversification, most of the members of this family are controlled by different regulatory circuits. If this was to be true, then the members of this gene family will exhibit different expression patterns under the same biotic or abiotic stress/stimuli. This is corroborated by studies in other plantsCitation17,Citation19 and it remains to be explored whether in jute other members of this gene family show a similar trend upon infection by M. phaseolina.
We speculate that when C. trilocularis comes in contact with M phaseolina, the expression of CtXTH1 increases with time, but its ortholog, CoXTH1 could possibly be under a different regulatory circuit and shows an opposite expression pattern. We cannot substantiate a differential regulation between the two genes as there are no studies on the hormonal regulation of jute XTH. Nevertheless, this study shows that two closely related XTH genes, with high homology in their sequences, show completely different expression patterns between fungus susceptible and resistant jute species, and it is highly possible that they are they are one of the major signaling components of jute M. phaseolina interaction.
Materials and methods
Fungal isolates and plant materials
M. phaseolina was cultured in PDA medium as described previously.Citation38 Seeds from both the susceptible (C. olitorious) and the resistant (C. trilocularis) plants were germinated on Petri dishes. These seedlings were allowed to grow on water only (without any growth medium) for three days in the absence of light. After the three days, two sets of seedlings from both species were sprayed with fungal suspension according to a protocol described earlier.Citation39,Citation40 A set of control (uninfected) seedlings from both species were collected during the first collection of infected seedlings. One set of infected seedlings from each species were collected at intervals of 15 and 24 h, from the time of fungal spray. These seedlings were washed with distilled water, snap-frozen in liquid nitrogen and stored at -80°C until further use.
RNA isolation and differential display
Total RNA was isolated from 1.0 g frozen tissue (seedlings) using Trizol reagent (Invitrogen) according to the manufacturer’s instructions. The concentration was measured spectrophotometrically. For differential display, first strand cDNA was prepared from all six samples using three anchored primers T12A, T12G and T12C. Three μg of total RNA was used in each 20µl reverse transcription (RT) reaction using SuperScriptIII RT (200 units/µl) (Invitrogen). One µl of anchored primers (50µM), 5 µl of RNA solution, 4 µl of 2.0 mM dNTPs mixture and 3 µl of RNase-free water were mixed in a thin walled PCR tube and heated to 65°C for 5 min and then, incubated on ice. Then 4 µl 5 × RT-buffer (Invirrogen), 1 µl 0.1 M DTT, 1 µl RNaseOUT (Invitrogen) and 1 µl SuperScriptIII RT (200 units/µl) (Invitrogen) were added in the tube and the mixture was incubated at 25°C for 5 min followed by 60 min at 50°C and heating at 70°C for 15 min. 0.5 µl of E. coli RNaseH (Invitrogen) was added and the mixture was further incubated at 37°C for 20 min. The use of three different anchored primers gave rise to three different samples for each species. One 11-mer arbitrary primer was used as 5′primer in different combinations with each of the anchored primers for PCR using a thermal cycler (Eppendorf Mastercycler Personal). The PCR conditions used are as following: 1 cycle at 94°C for 3 min and 40 cycles at 94°C for 40 sec, 40°C for 50 sec and 72°C for 2 min followed by a final extension at 72°C for 5 min. Ten μl of each reaction was loaded onto 10% polyacrylamide gel and electrophoresis was performed for 3 h at 70 V. Following separation, the amplified products were stained with silver nitrate using a method described earlier.Citation41 For a list of primers used in this article, please see .
Table 2. Primers used in this study
Differentially expressed cDNA of interest
The differentially expressed band was excised and incubated overnight in 2X volume (~100µL) of elution buffer (0.5M EDTA pH 8.0, 1M ammonium acetate) at 37°C with gentle shaking. The cDNA was ethanol precipitated and re-amplified using the arbitrary and the anchored primer (the primer pair that amplified the differentially expressed gene) and cloned into the pCR8/GW/TOPO TA Cloning vector (Invitrogen) following supplier’s protocol and propagated in E. coli (DH5α) cells in LB medium. The cloned fragment was sequenced using M13 forward and M13 reverse primers.
Primer based gene walking and rapid amplification of cDNA ends (RACE)
BLASTn and BLASTx search indicated the sequence to have high homology with an enzyme xyloglucan endotransglycosylase/hydrolase. A downstream degenerate primer XTHD2Rev was designed and checked by PCR (35 cycles, 95°C for 50 sec, 60°C for 40 sec, and 72°C for 1 min) in combination with gene specific primer XTHGF using genomic DNA as template. To explore the 3′ end of the differentially expressed gene, 3′ RACE was performed using the 3′ RACE system (Invitrogen) as per supplier’s instructions. cDNA was synthesized using adaptor primer AP. PCR of the cDNA was performed using gene specific forward primer XTH3′F and AUAP for 35 cycles (35 cycles, 95°C for 40 sec, 60°C for 30 sec, and 72°C for 1 min). For 5′ RACE, two gene specific primers were custom synthesized (GSP and GSP2). The first strand cDNA was prepared from mRNA using the primer GSP. For purification and the other subsequent steps, recommendations of the manufacturer were followed (5′ RACE system, Invitrogen). PCR of target cDNA was performed with GSP and AAP primers for 35 cycles (95°C for 1 min, 60°C for 40 sec, and 72°C for 1 min). Nested PCR of the first round PCR product was performed using GSP2 and AUAP for 40 cycles (95°C for 1 min, 60°C for 40 sec, and 72°C for 1 min). Another gene specific primer (XTH 3′R) was designed from the 3′ end and PCR amplification from the genomic DNA of C. trilocularis using XTH 3′F and XTH 3′R (35 cycles, 95°C for 1 min, 58°C for 40 sec, and 72°C for 50 sec) produced a 632bp product which led to identification of an intron. Interestingly, this same pair of primers yielded a PCR product of similar size from C. olitorious gDNA. Both the bands were sequenced and homology search for amplified C. olitorious gDNA band showed that it was also an XTH (CoXTH1) and the aforementioned approaches were also applied to determine its sequence. PCR amplification of gDNA of C. olitorious and C. trilocularis with XTH5′F and XTHGR primers (35 cycles, 95°C for 1 min, 59°C for 40 sec, and 72°C for 1 min) helped to identify the introns of CtXTH1 and CoXTH1.
Southern blot analysis
15μg of C. trilocularis and C. olitorious gDNA were digested with EcoRI (Invitrogen) and BamHI (Invitrogen) and transferred to a positively charged nylon membrane (Amersham Hybond™-N) according to manufacturer’s protocol. CtXTH1 and CoXTH1 sequences were ampli□ed using XTH5'F and XTHGR primers and labeled with digoxigenin-dUTP following manufacturer’s protocol (DIG nucleic acid detection and labeling kit, Roche). XTH specific sequences blotted on nylon membrane were detected following an immunologic assay according to manufacturer’s protocol ().
Semiquantitative reverse-transcription-PCR
The quantity and quality of six RNA samples () were measured both spectrophotometrically and by agarose gel electrophoresis. . First strand cDNA was synthesized using XTH 3′R primer. PCR amplification of the cDNA was performed for 30 cycles using primers XTH GF and XTH 3′R following the amplification conditions mentioned earlier. β-actin was used as a normalization control for reverse transcription polymerase chain reaction (RT-PCR).
Table 1. Different samples for differential gene expression study
Real-time PCR analysis
First strand cDNA synthesis (for six samples, two replicates each) was performed with 3µg of total RNA using XTH 3′R primer and Superscript III reverse transcriptase (Invitrogen) following the manufacturer’s recommendations. Real time quantifications were performed in Roche LightCycler® 480 Real-Time PCR System using LightCycler® 480 SYBR Green I Master kit (Roche) in a 20µL PCR volume containing 10µL of 2X LightCycler® 480 SYBR Green I Master (contains FastStart Taq DNA Polymerase, reaction buffer, dNTP mix, SYBR Green I dye, and MgCl2), 1.5 µL cDNA and 0.15 µM of each of the gene specific and β-actin gene primers (internal control). The thermal cycling parameters included a pre-incubation at 95°C for 5 min and 35 cycles of amplification (95°C for 40 sec, 58°C for 40 sec, and 72°C for 40 sec). Following the amplification, a three stage cycle was used for the determination of the melting curve (95°C for 0 sec, 70°C for 1 min and 95°C for 0 sec at continuous acquisition mode). The data was analyzed using the 2-ΔΔCt method.Citation42,Citation43
Additional material
Download Zip (90.9 KB)Acknowledgments
This research was supported by the US. Department of Agriculture (USDA) grant. We are thankful to BJRI authority for their support. We also thank M. Maksudul Alam (University of Texas at Dallas, USA) for help with the TA-cloning. Thanks are also due to Mahdi M Moosa for his valuable comments on the manuscript.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Ashraf H, Javaid A. Evaluation of antifungal activity of Meliaceae family against Macrophomina phaseolina.. Mycopath 2007; 5:81 - 4
- Bhowal. J, Ghosh. S, Guha. AK, Chatterjee BP. Infection of jute seedlings by the phytopathogenic fungus Macrophomina phaseolina mediated by endogenous lectin. Research Journal of Microbiology 2006; 1:51 - 60; http://dx.doi.org/10.3923/jm.2006.51.60
- Indera K, Singh T, Machado CC, Sinclair JB. Histopathology of soybean seed infection by Macrophomina phaseolina.. Phytopathology 1986; 76:532 - 5; http://dx.doi.org/10.1094/Phyto-76-532
- Khan SN. Macrophomina phaseolina as causal agent for charcoal rot of sunflower. Mycopath 2007; 5:111 - 8
- Yang SM, Owen DF. Symptomatology and Detection of Macrophomina phaseolina in Sunflower Plants Parasitized by Cylindrocopturus adspersus Larvae. Phytopathology 1982; 72:819 - 21; http://dx.doi.org/10.1094/Phyto-72-819
- Biswas AC, Kabir MQ, Ahmed QA. Differential response of wild species of jute to stem rot (Macrophomina phaseoli). Agr Pakistan 1968; 11:165 - 7
- Liang P, Pardee AB. Differential display of eukaryotic messenger RNA by means of the polymerase chain reaction. Science 1992; 257:967 - 71; http://dx.doi.org/10.1126/science.1354393; PMID: 1354393
- Hückelhoven R. Cell wall-associated mechanisms of disease resistance and susceptibility. Annu Rev Phytopathol 2007; 45:101 - 27; http://dx.doi.org/10.1146/annurev.phyto.45.062806.094325; PMID: 17352660
- Nishitani K, Tominaga R. In vitro molecular weight increase in xyloglucans by an apoplastic enzyme preparation from epicotyls of Vigna angularis.. Physiol Plant 1991; 82:490 - 7; http://dx.doi.org/10.1111/j.1399-3054.1991.tb02937.x
- Okazawa K, Sato Y, Nakagawa T, Asada K, Kato I, Tomita E, et al. Molecular cloning and cDNA sequencing of endoxyloglucan transferase, a novel class of glycosyltransferase that mediates molecular grafting between matrix polysaccharides in plant cell walls. J Biol Chem 1993; 268:25364 - 8; PMID: 8244968
- Sulová Z, Baran R, Farkaš V. Release of complexed xyloglucan endotransglycosylase (XET) from plant cell walls by a transglycosylation reaction with xyloglucan-derived oligosaccharides. Plant Physiol Biochem 2001; 39:927 - 32; http://dx.doi.org/10.1016/S0981-9428(01)01313-4
- Bauer WD, Talmadge KW, Keegstra K, Albersheim P. The Structure of Plant Cell Walls: II. The Hemicellulose of the Walls of Suspension-cultured Sycamore Cells. Plant Physiol 1973; 51:174 - 87; http://dx.doi.org/10.1104/pp.51.1.174; PMID: 16658281
- Hayashi T. Xyloglucans in the Primary Cell Wall. Annu Rev Plant Physiol Plant Mol Biol 1989; 40:139 - 68; http://dx.doi.org/10.1146/annurev.pp.40.060189.001035
- Rose JKC, Braam J, Fry SC, Nishitani K. The XTH family of enzymes involved in xyloglucan endotransglucosylation and endohydrolysis: current perspectives and a new unifying nomenclature. Plant Cell Physiol 2002; 43:1421 - 35; http://dx.doi.org/10.1093/pcp/pcf171; PMID: 12514239
- Carpita NC, Gibeaut DM. Structural models of primary cell walls in flowering plants: consistency of molecular structure with the physical properties of the walls during growth. Plant J 1993; 3:1 - 30; http://dx.doi.org/10.1111/j.1365-313X.1993.tb00007.x; PMID: 8401598
- Nishitani K. The Role of Endoxyloglucan Transferase in the Organization of Plant Cell Walls. In: Kwang WJ, ed. International Review of Cytology: Academic Press, 1997:157-206.
- Xu W, Campbell P, Vargheese AK, Braam J. The Arabidopsis XET-related gene family: environmental and hormonal regulation of expression. Plant J 1996; 9:879 - 89; http://dx.doi.org/10.1046/j.1365-313X.1996.9060879.x; PMID: 8696366
- Bendtsen JD, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: SignalP 3.0. J Mol Biol 2004; 340:783 - 95; http://dx.doi.org/10.1016/j.jmb.2004.05.028; PMID: 15223320
- Michailidis G, Argiriou A, Darzentas N, Tsaftaris A. Analysis of xyloglucan endotransglycosylase/hydrolase (XTH) genes from allotetraploid (Gossypium hirsutum) cotton and its diploid progenitors expressed during fiber elongation. J Plant Physiol 2009; 166:403 - 16; http://dx.doi.org/10.1016/j.jplph.2008.06.013; PMID: 18789555
- Baumann MJ, Eklöf JM, Michel G, Kallas AM, Teeri TT, Czjzek M, et al. Structural evidence for the evolution of xyloglucanase activity from xyloglucan endo-transglycosylases: biological implications for cell wall metabolism. Plant Cell 2007; 19:1947 - 63; http://dx.doi.org/10.1105/tpc.107.051391; PMID: 17557806
- Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 1997; 18:2714 - 23; http://dx.doi.org/10.1002/elps.1150181505; PMID: 9504803
- Johansson P, Brumer H 3rd, Baumann MJ, Kallas AM, Henriksson H, Denman SE, et al. Crystal structures of a poplar xyloglucan endotransglycosylase reveal details of transglycosylation acceptor binding. Plant Cell 2004; 16:874 - 86; http://dx.doi.org/10.1105/tpc.020065; PMID: 15020748
- Liang P, Pardee AB. Recent advances in differential display. Curr Opin Immunol 1995; 7:274 - 80; http://dx.doi.org/10.1016/0952-7915(95)80015-8; PMID: 7546389
- Yokoyama R, Nishitani K. A comprehensive expression analysis of all members of a gene family encoding cell-wall enzymes allowed us to predict cis-regulatory regions involved in cell-wall construction in specific organs of Arabidopsis. Plant Cell Physiol 2001; 42:1025 - 33; http://dx.doi.org/10.1093/pcp/pce154; PMID: 11673616
- Albert M, Werner M, Proksch P, Fry SC, Kaldenhoff R. The cell wall-modifying xyloglucan endotransglycosylase/hydrolase LeXTH1 is expressed during the defence reaction of tomato against the plant parasite Cuscuta reflexa. Plant Biol (Stuttg) 2004; 6:402 - 7; http://dx.doi.org/10.1055/s-2004-817959; PMID: 15248122
- Yokoyama R, Nishitani K. Functional Diversity of Xyloglucan-Related Proteins and its Implications in the Cell Wall Dynamics in Plants. Plant Biol 2000; 2:598 - 604; http://dx.doi.org/10.1055/s-2000-16643
- Smith RC, Fry SC. Endotransglycosylation of xyloglucans in plant cell suspension cultures. Biochem J 1991; 279:529 - 35; PMID: 1953647
- Nishitani K. Construction and restructuring of the cellulose-xyloglucan framework in the apoplast as mediated by the xyloglucan-related protein family—A hypothetical scheme. J Plant Res 1998; 111:159 - 66; http://dx.doi.org/10.1007/BF02507162
- Thompson JE, Smith RC, Fry SC. Xyloglucan undergoes interpolymeric transglycosylation during binding to the plant cell wall in vivo: evidence from 13C/3H dual labelling and isopycnic centrifugation in caesium trifluoroacetate. Biochem J 1997; 327:699 - 708; PMID: 9581545
- Takeda T, Furuta Y, Awano T, Mizuno K, Mitsuishi Y, Hayashi T. Suppression and acceleration of cell elongation by integration of xyloglucans in pea stem segments. Proc Natl Acad Sci U S A 2002; 99:9055 - 60; http://dx.doi.org/10.1073/pnas.132080299; PMID: 12084943
- Maldonado-Mendoza IE, Dewbre GR, Blaylock L, Harrison MJ. Expression of a xyloglucan endotransglucosylase/hydrolase gene, Mt-XTH1, from Medicago truncatula is induced systemically in mycorrhizal roots. Gene 2005; 345:191 - 7; http://dx.doi.org/10.1016/j.gene.2004.10.028; PMID: 15716119
- Potter I, Fry SC. Xyloglucan endotransglycosylase activity in pea internodes. Effects of applied gibberellic acid. Plant Physiol 1993; 103:235 - 41; http://dx.doi.org/10.1104/pp.103.1.235; PMID: 8208849
- Braam J, Davis RW. Rain-, wind-, and touch-induced expression of calmodulin and calmodulin-related genes in Arabidopsis. Cell 1990; 60:357 - 64; http://dx.doi.org/10.1016/0092-8674(90)90587-5; PMID: 2302732
- Peschke VM, Sachs MM. Characterization and expression of transcripts induced by oxygen deprivation in maize (Zea mays L.). Plant Physiol 1994; 104:387 - 94; http://dx.doi.org/10.1104/pp.104.2.387; PMID: 7909162
- Burstin J. Differential expression of two barley XET-related genes during coleoptile growth. J Exp Bot 2000; 51:847 - 52; http://dx.doi.org/10.1093/jexbot/51.346.847; PMID: 10948210
- Antosiewicz DM, Purugganan MM, Polisensky DH, Braam J. Cellular localization of Arabidopsis xyloglucan endotransglycosylase-related proteins during development and after wind stimulation. Plant Physiol 1997; 115:1319 - 28; http://dx.doi.org/10.1104/pp.115.4.1319; PMID: 9414546
- Vissenberg K, Martinez-Vilchez IM, Verbelen JP, Miller JG, Fry SC. In vivo colocalization of xyloglucan endotransglycosylase activity and its donor substrate in the elongation zone of Arabidopsis roots. Plant Cell 2000; 12:1229 - 37; PMID: 10899986
- Aboshosha SS, Alla SIA, El-Korany AE, El-Argawy E. Characterization of Macrophomina phaseolina isolates affecting sunflower growth in El-Behera Governorate, Egypt. International Journal of Agriculture and Biology 2007; 9:807 - 15
- Alam M, Sharmin S, Nabi Z, Mondal S, Islam M, Nayeem S, et al. A Putative Leucine-Rich Repeat Receptor-Like Kinase of Jute Involved in Stress Response. Plant Mol Biol Rep 2010; 28:394 - 402; http://dx.doi.org/10.1007/s11105-009-0166-4
- Mahmood N, Azam MS, Islam MS, Sharmin S, Moosa MM, Hossain MA, et al. Differentially expressed transcripts of wild and cultivated jute (Corchorus spp.) varieties upon fungal (Macrophomina phaseolina) infection. Annals of Biological Research 2010; 1:120 - 7
- Qu L, Li X, Wu G, Yang N. Efficient and sensitive method of DNA silver staining in polyacrylamide gels. Electrophoresis 2005; 26:99 - 101; http://dx.doi.org/10.1002/elps.200406177; PMID: 15624131
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Δ Δ C(T)) Method. Methods 2001; 25:402 - 8; http://dx.doi.org/10.1006/meth.2001.1262; PMID: 11846609
- Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 2008; 3:1101 - 8; http://dx.doi.org/10.1038/nprot.2008.73; PMID: 18546601