Abstract
Mini chromosome maintenance (MCM) proteins 2–7, a subgroup of the large AAA ATPase family are critically required for eukaryotic DNA replication. These proteins are most likely responsible for unwinding DNA at the replication forks. Besides this function, some MCMs are also involved in other chromosome transactions such as transcription, chromatin remodeling and genome stability. All the MCMs contain a conserved region of ~200 amino acids responsible for nucleotide binding. The importance of MCM proteins is evident by the fact that deregulation of the activity of MCM family of proteins appears to be directly linked to human carcinogenesis. This article will focus on members of this important family of proteins from the malaria parasite Plasmodium falciparum and their comparison with the human host.
Introduction
The minichromosome maintenance (MCM) genes were first identified in the yeast Saccharomyces cerevisiae by a genetic screen for minichromosome maintenance-defective mutants.Citation1 The mutants defective in each of these genes exhibit high rates of mitotic chromosome loss and recombination. MCM4 (CDC54) and MCM7 (CDC47) were isolated as cell cycle division mutants and MCM6,Citation2 as a chromosome segregation mutant (mis5) was originally isolated in Schizosaccharomyces pombe.Citation3 The MCM proteins are essential replication initiation factors consisting of six sequence-related subunits MCM2 to 7, which are evolutionally conserved in all eukaryotes and archaea. Because of their involvement in a fundamental cellular process, all six of these genes are essential for viabilityCitation4,Citation5 and renamed MCM2 through MCM7,Citation6 the name MCM1 had already been categorized as a transcription factor and it does not belong to the MCM2–9 family.Citation7 It has been suggested that MCM family in human is constituted of eight members, falling into two different groups, one constituted by the MCM2–7 complex and the other by MCM8 and MCM9, which are present only in higher eukaryotes. Each MCM protein is essential for DNA replication because deletion of any one of the six subunits results in cell death in yeast. The six eukaryotic MCM proteins share significant sequence similarity with one another, concentrating on a nearly 250- amino-acid region that encodes the ATPase active site (AAA domain).Citation8 MCMs drive the formation of prereplicative complexes (PRCs) during the G1 phase of the cell cycle. It has been well established that the conversion from the cell cycle to G0 phase (quiescence) is due to the downregulation of the MCM2–7 protein complexes.
AAA or AAA+ is an abbreviation for ATPases associated with diverse cellular activities. They have a common conserved module of approximately 230–250 amino acid residues. They contain ring-shaped P-loop NTPases, which exert their activity through the energy-dependent remodeling or translocation of macromolecules.Citation9,Citation10 MCM family is a large and functionally diverse protein family belonging to the AAA+ superfamily. AAA proteins form ATPase active sites at clefts between two subdomains: one containing a series of parallel β-strands (P loop) and a second called the lid.Citation11 Both subdomains contain conserved active-site motifs. The P loop contains motifs involved in binding ATP (Walker A box) and positioning the nucleophilic water molecule (Walker B box and sensor 1), while the lid domain contains motifs that contact the γ-phosphate of ATP (arginine finger and sensor 2).Citation6 Three indispensable motifs known as Walker A or motif I, Walker B or motif II and arginine finger are characteristic of all of the 8 MCM proteins.
During the G1 phase of the cell cycle, MCM2–7 is carried to origins of replication in an inactive state by Cdt1 to form the prereplication complex (pre-RC),Citation12 a well ordered assembly that contains the AAA+ proteins Cdc6 and the hetero-hexameric origin recognition complex (Orc1–6).Citation12,Citation13 All the six MCM subunits 2 to 7 co-localize to origins of replication during pre-RC formation.Citation13-Citation15 The inactivation or loss of any of the six MCM subunits during G1 phase inhibits pre-RC formation in vivo in yeast.Citation16 The loading of MCM2–7 onto DNA requires ATP hydrolysis by both Orc1–6 and Cdc6.Citation17,Citation18 Once the loading has been achieved, Orc1–6 and Cdc6 are no more required for MCM2–7 retention at the originCitation17,Citation19 and they are unnecessary for subsequent DNA replication.Citation20,Citation21
MCM proteins are related to cell proliferation therefore they serve as useful biomarkers for cancer screening, surveillance and prognosis. The deregulation of MCM expression disrupts genetic stability and the overexpression of MCM subunits has been reported in various premalignant dysplastic lesions and cancers.Citation22 It has been reported that MCM2, MCM3 and MCM7 expression are frequently deregulated in medulloblastoma (MB).Citation23 MCM family of proteins was reported to be upregulated in meningiomas and it was suggested that these can be used as diagnostic markers. Significant increase in expression in meningiomas was reported for MCM2 (8 fold), MCM3 (5 fold), MCM4 (4 fold), MCM5 (4 fold), MCM6 (3 fold), and MCM7 (5 fold).Citation24 Although the roles of many proteins involved in the initiation of replication are understood, the role of MCM10 remains controversial.Citation25,Citation26 A key process of replication initiation is to convert inactive MCM2–7 to active Cdc45-MCM-GINS (CMG) replicative helicase. MCM10 is supposed to have an essential role in functioning of the CMG replicative helicase independent of assembly of a stable CMG complex at origins.Citation26
MCM proteins have been isolated from a number of plants but their straightforward role is yet to be established.Citation27 Their abundant presence in plants is still ambiguous. It has been suggested that other than their established role in DNA unwinding, they might have the additional roles most likely in stress tolerance.Citation27 It has been reported recently that pea MCM6 transcript is upregulated in pea plant in response to high salinity and cold stress but not with ABA, drought and heat stress. The overexpression of pea MCM6 single subunit in tobacco plant promotes salinity stress tolerance without affecting its yield.Citation28
Basic understanding of the molecular basis of cell growth and differentiation in the malaria parasite is essential for the development of novel chemotherapeutic agents. The malaria parasite has a complex life cycle in mosquito and human hosts.Citation29 There are five points in the life cycle of the parasite where cell division/replication occurs.Citation30,Citation31 The MCM complex is recruited to the pre-replication complex (pre-RC) before DNA synthesis. Although the parasite has a tight control mechanism for cell proliferation and differentiation during various points of its life cycle, however very little is known about the molecular machinery that regulates this process. Different studies have reported the involvement of only ORC1 and MCM4 in pre-replication formation during replication and both have been reported to be expressed only in gametocytes.Citation32,Citation33 However presence of all the six subunits of the MCM complex in the P. falciparum genome has been reported.Citation33 In the present manuscript we present a comprehensive in silico analysis of MCM family of proteins from P. falciparum and their comparison with human host.
Results and Discussion
The genome of P. falciparum, available at www.plasmodb.org was investigated using ‘MCM’ as query. The results of this search presented in showed a total of 8 hits. The PlasmoDB number for MCM2–9 and other details are listed in . All the 8 MCM members from this list were used to BLAST with H. sapiens and the comparative analysis are presented under different categories in the following sections. MCM proteins show considerable sequence conservation particularly in a 200 amino acid residue domain, which is located almost in the center of these large proteins. Walker A motif contains the consensus sequence [(G)xxxxGK[S/T], where x is any residue and lysine residue is present which is characteristic of all the ATP-binding proteins. It has been observed that the Walker A motif sequence in all the MCMs is slightly deviated from the consensus and the glycines in the motif GK(S/T) are substituted by serine or alanine.Citation8 The Walker B motif (hhhh[D/E] has conserved nucleotide phosphate-binding motif (where h is a hydrophobic residue). These are the characteristic feature of members of the P-loop NTPase domain superfamily. The Walker A motif binds the β-γ phosphate moiety of the bound nucleotide (typically ATP or GTP) and Walker B motif binds with the Mg2+ cation.Citation34 All the MCMs contain a Zn finger and another short motif SRFD, which is present approximately 70 residues after the Walker B motif and defines an arginine finger motif.
Table 1. MCM family of Plasmodium falciparum
MCM2
It has been well established that HsMCM2 contributes to a variety of nuclear functions in addition to DNA replication. The detailed biochemical characterization of HsMCM2 showed that the C-terminal region of HsMCM2 contains ssDNA- binding activity that inhibits the DNA helicase activity.Citation35 On the other hand using pulldown analysis it was reported that two fragments from the central region were mainly responsible for the interaction between HsMCM2 and HsMCM4.Citation35
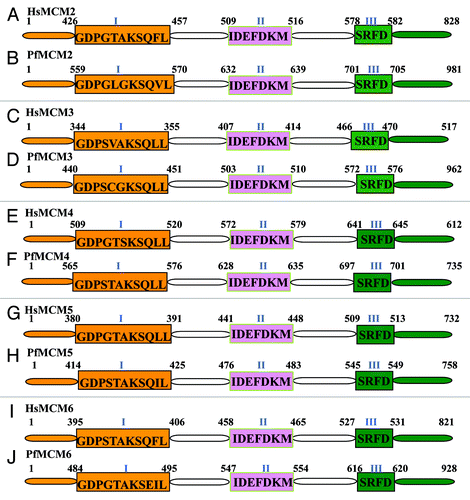
The gene with PlasmoDB number PF14_0177 is a homolog of human MCM2 and is located on chromosome 14. PfMCM2 protein is slightly larger in size as compared with its human homolog (). It contains additional 163 amino acids in comparison to the human counterpart. The difference in size mainly lies in the N-terminal extension ( and B). The sequence of all the domains and the comparison with the human counterpart shows that Walker A motif of PfMCM2 follows the consensus sequence but in human counterpart A is present instead of G ( and B). In motif II i.e., Walker B motif PfMCM2 and HsMCM2 obey the consensus sequence, although there is little sequence dissimilarity ( and B). Similar to other eukaryotic MCMs, PfMCM2 also contains a putative C4-type zinc finger domain at its N-terminal region. The zinc finger domain may have a role in the binding of PfMCMs to chromatin because these domains are known to be responsible for protein-DNA and protein-protein interactions and therefore contribute to complex assembly. The arginine finger motif of MCM2 of P. falciparum and human are highly conserved ( and B). The expression of PfMCM2 predominantly in late trophozoites and during schizont maturation,Citation32 concur with DNA replication in Plasmodium.Citation33
Figure 1. Comparison of MCM2–6 of Plasmodium falciparum with their human homologs. All the sequence data used in the analysis were downloaded from PlasmoDB (www.plasmodb.org). The downloaded sequences were used as query to match with the human homolog using BLAST search (www.ncbi.nlm.nih.gov). The corresponding human sequence was retrieved and the domain analysis was done. The results were further checked manually to confirm that all the domains are present and then used in the figures. Similarly the P. falciparum sequence was also analyzed and the results are presented in figures. The conserved sequences of each domain are written inside the boxes. The text in blue refers to the names of various conserved domains and the numbers refer to the amino acids positions of various domains. The number I, II and III refer to Walker A or motif I, Walker B or motif II and arginine finger motif respectively. This figure is not drawn to scale.
MCM3
The phosphorylation of HsMCM3 has been well studied. It was reported that cyclin B–CDK1 catalyzes phosphorylation of HsMCM3 at Ser-112, thereby regulating HsMCM3 association with other HsMCM2–7 subunits and loading of HsMCM3 onto chromatin.Citation36 In a recent study it has been reported that MCM3 is a substrate of cyclin E/Cdk2 and can be phosphorylated by cyclin E/Cdk2 at Thr-722.Citation37 The HsMCM3 T722A mutant binds chromatin much less efficiently as compared with wild type HsMCM3 suggesting that this phosphorylation site is involved in HsMCM3 loading onto chromatin. The knockdown of HsMCM3 does not affect the S phase entry and progression, indicating that a small fraction of HsMCM3 is sufficient for normal S phase completion.Citation37 These studies strongly suggest that HsMCM3 phosphorylation is a critical posttranslational modification that regulates assembly and activity of the HsMCM2–7 complex.
The gene with PlasmoDB number PFE1345c is a homolog of human MCM3 and is located on chromosome 5 and is slightly larger in size as compared with its human homolog (). It has additional 154 amino acids; ~100 at N-terminal and 50 amino acids at its C-terminal region respectively ( and D). The sequence comparison of all the domains with human counterpart indicates that except Walker A motif (motif I) all the other domains are identical ( and D).
MCM4
The importance of phosphorylation for the activity of HsMCM4 has been well studied. The phosphorylation at sites 3 and 32 of HsMCM4 required CDK2 in HeLa cells and this phosphorylated HsMCM4 had several distinct and site-specific roles in MCM function.Citation38 It was reported that the central region of HsMCM2, which contains zinc finger and ATPase motif interacts with HsMCM4.Citation35 In a recent study the first human mutation in MCM4 gene has been reported and it has been shown to be associated with adrenal insufficiency, short stature, and natural killer (NK) cell deficiency.Citation39 In a related study it was reported that the partial HsMCM4 deficiency results in a genetic syndrome of growth retardation with adrenal insufficiency and selective NK deficiency.Citation40
The gene with PlasmoDB number PF13_0095 is a homolog of human MCM4 and is located on chromosome 13 and is larger in size as compared with its human homolog (). It contains 197 additional amino acids in comparison to human homolog. The N-terminal and C-terminal of PfMCM4 has 56 and 38 amino acid more than the human counterpart ( and F). The conserved motif of PfMCM4 showed difference in sequences in Walker A motif (motif I) while Walker B (motif II) and arginine finger motifs (motif III) are identical to HsMCM4 ( and F). PfMCM4 contains the zinc finger domain at the N-terminal region. This domain may have a role in the protein-DNA interactions. In the case of Walker A motif, the consensus sequence i.e., GXXXXGK(S/T) is not followed by both PfMCM4 and HsMCM4 ( and F). In a previous study it has been reported that PfMCM4 contains some unique features such as it is the largest of all the MCM4 and contains insertions at few places within its entire sequence including the zinc finger domain.Citation31 It was also reported that PfMCM4 is expressed specifically in the sexual erythrocytic stages indicating that PfMCM4 may be involved in gametogenesis where DNA is replicated.Citation31
MCM5
Cyclin E has been shown to directly interact with and colocalize on centrosomes with HsMCM5 in a centrosomal localization sequence (CLS)-dependent but Cdk2-independent manner.Citation41 The HsMCM5 domain responsible for interaction with cyclin E is reported to be highly conserved in MCM5 proteins from yeast to mammals. The expression of HsMCM5 or its cyclin E-interacting domain significantly inhibits over-duplication of centrosomes indicating that proteins involved in DNA replication might also regulate centrosome duplication.Citation41 In a related study it was reported that cyclins E and A sequentially prevent centrosome reduplication throughout interphase by recruitment of HsMCM5 and Orc1.Citation42 HsMCM-5 expression has been reported to be associated with clinicopathological parameters in gastric adenocarcinoma.Citation43
The gene with PlasmoDB number PFL0580w is a homolog of HsMCM5. PfMCM5 is almost similar in size and is located on chromosome number 12 (). The comparative analysis of their motifs showed that only Walker A motifs (motif I) are identical and other two motifs have little dissimilarity in their sequences ( and H).
MCM6
MCM proteins are essential for DNA replication and MCM6 is one subunit of HsMCM2–7 complex that serves as the replicative helicase in DNA replication. It is well established that Cdt1 physically interacts with the MCM complex and this interaction mainly occurs between Cdt1 and MCM6 in human cells. The detailed analysis indicated that the C-terminal 79 residues of hCdt1 interact with the C-terminal 113 residues of HsMCM6 while the large N-terminal Orc6-binding domain recruits Cdt1/MCM2–7 to ORC complex.Citation44 HsMCM6 has been suggested to be a suitable marker for evaluation of the proliferation potential of craniopharyngioma. The study of expression of HsMCM6 and its correlation with recurrence of the tumor in craniopharyngiomas suggested no difference in HsMCM6 expression between the primary and recurring tumors.Citation45
The gene with PlasmoDB number PF13_0291 is a homolog of HsMCM6. PfMCM6 is larger by 108 amino acids (). The difference in size lies in the N-terminal region of PfMCM6 ( and J). The comparative analysis of the conserved motifs showed slight differences in their amino acid sequences in all three motifs ( and J). The zinc finger domain in PfMCM6 is also present at its N-terminal region and may play a role in the binding of PfMCM6 to chromatin. Walker A motifs of both MCM6 do not obey the consensus sequences ( and J). In a previous study the expression of PfMCM6 was observed predominantly in late trophozoites and during schizont maturation.Citation33 During the trophozoite stage, the antibody against PfMCM6 pulls down PfMCM2 very inefficiently.Citation33 It was also reported that only PfMCM6 is associated with the chromatin fraction at all stages of growth and PfMCM6 is also present in both the nucleosolic and cytoplasmic fractions.Citation33 It might be possible that the PfMCM6 subunit makes direct contact with chromatin while other subunits are associated through protein–protein interactions.
MCM7
Recent studies suggest that in a variety of human malignancies MCM7 is amplified and overexpressed. It has been reported that dysregulation of MCM7 expression is observed in various types of cancer and it correlates with a negative outcome in patients with non-small cell lung cancer after surgical resection. Therefore MCM7 can be explored as a potential therapeutic and prognostic target in lung and other cancers.Citation46
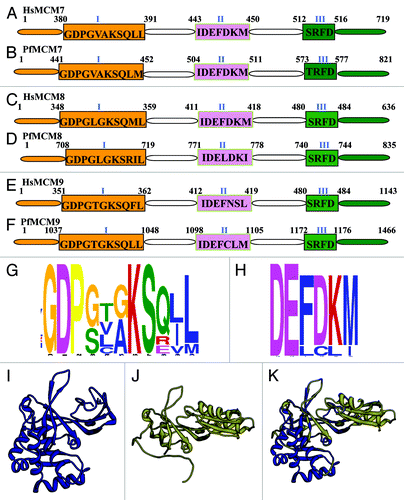
The gene with PlasmoDB number PF07_0023 is a homolog of HsMCM7 and it is located on chromosome 7 (). PfMCM7 contains additional 102 amino acids as compared with its human counterpart. These amino acids are distributed on both the N-terminal and C-terminal sides of the protein ( and B). Walker A motifs (motif I) in both Pf and HsMCM7 do not agree with the consensus sequences ( and B). The comparison of HsMCM7 with PfMCM7 indicates that Walker B motifs (motif II) are identical but the arginine finger motif (motif III) displays variation in one amino acid ( and B). Similar to other MCMs, PfMCM7 contains putative C4-type zinc finger domain at its N-terminal region. This domain probably has a role in the binding of PfMCM7 to DNA. It has been shown that during the trophozoite stage, the antibody against PfMCM6 does not pull-down PfMCM7.Citation33 The expression of PfMCM7 polypeptides was predominantly observed in late trophozoites and during schizont maturationCitation33,Citation47 and decreased in the ring stages, which is in agreement with DNA replication in Plasmodium. It was also reported that PfMCM7 is found in the nucleosolic and cytoplasmic fractions throughout the intraerythrocytic life cycle and increased in both fractions from ring to schizont stages.Citation33
Figure 2. (A-F) Comparison of MCM7–9 of Plasmodium falciparum with their human homologs. All the sequence data used in the analysis were downloaded from PlasmoDB (www.plasmodb.org). The downloaded sequences were used as query to match with the human homolog using BLAST search (www.ncbi.nlm.nih.gov). The corresponding human sequence was retrieved and the domain analysis was done. Other details are as in . This figure is not drawn to scale. (G and H) Distribution of amino acids at various positions in (G). ‘Walker motif A; (H) ‘Walker motif B’ of MCM2–9 of P. falciparum. Single letter code for amino acids has been used. This analysis was done using MEME program. (I-K) The modeled structure of (I). template (J). PfMCM9 and (K). superimposition are shown. The PfMCM9 sequence was submitted to Swissmodel server and the structure was obtained. The molecular graphic images were produced using the UCSF Chimera package from the resource for Biocomputing, Visualization, and Informatics (www.cgl.ucsf.edu/chimera) at the University of California, San Francisco (supported by NIH P41 RR-01081). The details are described in text.
MCM8
MCM8 is a new evolutionarily conserved family member but its homolog is not present in yeast.Citation48 It may function alone or with other family members in DNA metabolism. HsMCM8 mRNA accumulates during the G1/S to G2/M phase, while its protein product is detectable throughout the normal cell cycle.Citation49 MCM8 is a crucial component of the pre-RC and the interaction between HsMCM8 and hCDC6 is required for pre-RC assembly.Citation49 In this study MCM8 appears to participate in pre-RC formation in human cells, although this is not the case in other systems. It has been reported that MCM8, like MCM7, colocalizes on a specific DNA segment of the c-Myc replication initiation zone (c-Myc replicator) with Cdc6.Citation50 The association of both MCM proteins with Cdc6 continues even after DNA replication is complete. It was reported that MCM8 may play a role in elongation and was strongly suggested that MCM8 functions with other known replication proteins in processes which accompany initiation of DNA replication.Citation50
The gene with PlasmoDB number PFL0560c is a homolog of HsMCM8 and PfMCM8 is located on chromosome 12 (). There is a large difference in their size; PfMCM8 is larger by 400 amino acids in comparison to HsMCM8. The difference in size is mainly attributable to a longer N-terminal region in PfMCM8 ( and D). The comparative analysis of the conserved motifs showed that only arginine finger motif (motif III) is highly conserved and identical in HsMCM8 and PfMCM8 but there are some differences in Walker A (motif I) and Walker B motifs (motif II) of HsMCM8 and PfMCM8 ( and D).
MCM9
HsMCM9 is a novel member of MCM family.Citation51,Citation52 Similar to HsMCM8, HsMCM9 is only present in the genome of higher eukaryotes. It showed 24–31% total amino acid identity with HsMCM2–MCM8 proteins and contains a unique C-terminal domain which has only weak homology to MCM2–7 and MCM8 but is conserved within MCM9 homologs.Citation53 The phylogenetic analysis on MCM family members indicated that MCM9 was most closely related to MCM8.Citation52 Further studies reported that HsMCM9 is expressed and required for loading the HsMCM2–7 helicase onto chromatin during pre-RC formation.Citation53
PFD0790c is the PlasmoDB number of the homolog of HsMCM9 (). PfMCM9 has additional 322 amino acids as compared with HsMCM9. PfMCM9 has extra 685 amino acid at its N-terminal while its C-terminal is shorter by 321 amino acid as compared with HsMCM9 ( and F). The motif comparison showed that only the arginine finger motif (motif III) is identical in HsMCM9 and PfMCM9 but there are some variations in Walker A (motif I) and Walker B (motif II) motifs of HsMCM9 and PfMCM9 ( and F).
To further analyze the conserved motifs in P. falciparum protein family, MEME Suite-GLAM2 version 4.8.0 was used. This analysis indicated that in P. falciparum MCM family in Walker A motif (motif I), 5 out of 11 amino acids are highly conserved and others are variable to some extent (). On the other hand in Walker B motif (motif II), two out of six amino acids are highly conserved and others are slightly variable ().
For structural modeling the sequence of PfMCM9 was submitted to the Swissmodel homology-modeling server (swissmodel.expasy.org/). PfMCM9 primary sequence residues 405 to 650 showed only:16.4% sequence identity to a MCM protein homolog from archaeal Methanobacterium thermoautotrophicum.Citation54 The structural modeling of the PfMCM9 was therefore done using the known crystal structure of this homolog as the template (PDB number 1LTL at www.rcsb.org). The ribbon diagram of the template is shown in and the predicted structure of PfMCM9 is shown in respectively. When the modeled structure of PfMCM9 was superimposed, it is clear that these structures superimpose partially (). Molecular graphic images were produced using the UCSF Chimera package (www.cgl.ucsf.edu/chimera) from the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco (supported by NIH P41 RR-01081).Citation55
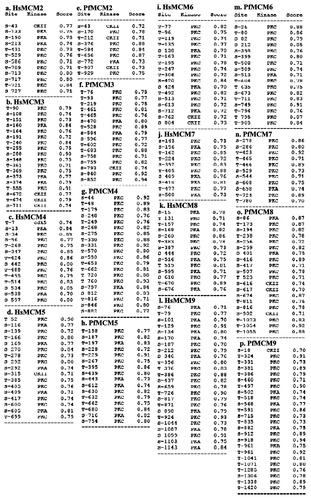
It is well established that post translational modification especially phosphorylation plays a very important role in regulating the function of various MCMs. The phosphorylation of MCM2, MCM3, MCM4, MCM6 and MCM7 has been observed in vivo and in vitro in different eukaryotic cellsCitation56,Citation57 however the same is not reported in case of PfMCMs.Citation33 Therefore the phosphorylation potential of all the HsMCMs and PfMCMs was analyzed using NetphosK at www.cbs.dtu.dk/services/NetPhosK.Citation58 The results of this analysis suggested that all of these proteins are prone to phosphorylation and contain the recognition sites mostly for PKC, followed by PKA, CKII and only few contain the sites for phosphorylation by PKB or cdk5 (). This analysis suggested that although there is a large variation in the size of PfMCMs and HsMCMs, the phosphorylation sites are almost equal in Hs and PfMCM3, MCM5, MCM6, MCM7 and MCM9 ( and f, d and h, i and m, j and n, l and p respectively). Furthermore it is interesting to note that PfMCM2 contains only 9 sites as opposed to HsMCM2, which contains 12 sites ( and e) and PfMCM4 and PfMCM8 each contain 20 phosphorylation sites as compared with HsMCM4 and HsMCM8, which contain only 14 sites each ( and g, k and o respectively). Overall in this manuscript, we have presented a comparative systematic analysis of MCM family from P. falciparum and human. These studies pave the way for further studies on this important family of proteins.
Figure 3. Phosphorylation sites in Hs and PfMCMs. The sequence of each MCM was submitted to www.cbs.dtu.dk/services/NetPhosK for predictions of kinase specific eukaryotic protein phosphorylation sites. A threshold of 0.7 was used and the sites predicted for each MCM are presented in the figure. CKII- casein kinase II, PKA-protein kinase A, PKB-protein kinase B, PKC-protein kinase C.
Materials and Methods
All the sequence data used in the analysis were downloaded from Plasmodium genome database available at www.plasmodb.org. The downloaded sequences were used as query and then matched with the human homolog using BLAST search (www.ncbi.nlm.nih.gov). The corresponding human sequences were retrieved and then their conserved domains were searched by using online CD search tool available on www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi. All the motifs in conserved domains were manually assigned. Similarly the motifs were manually assigned in P. falciparum sequence and the data are presented in figures. To identify conserved motifs within MCM family of P. falciparum, MEME Suite-GLAM2 version 4.8.0 was employed (meme.nbcr.net/meme/intro.html) using default settings.Citation59
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
The work in R.T's laboratory is partially supported by the Department of Biotechnology, Department of Science and Technology and Defense Research and Development Organization grants. Infra-structural support from the Department of Biotechnology, Government of India is gratefully acknowledged.
References
- Maine GT, Sinha P, Tye BK. Mutants of S. cerevisiae defective in the maintenance of minichromosomes. Genetics 1984; 106:365 - 85; PMID: 6323245
- Hennessy KM, Lee A, Chen E, Botstein D. A group of interacting yeast DNA replication genes. Genes Dev 1991; 5:958 - 69; http://dx.doi.org/10.1101/gad.5.6.958; PMID: 2044962
- Takahashi K, Yamada H, Yanagida M. Fission yeast minichromosome loss mutants mis cause lethal aneuploidy and replication abnormality. Mol Biol Cell 1994; 5:1145 - 58; PMID: 7865880
- Gómez EB, Catlett MG, Forsburg SL. Different phenotypes in vivo are associated with ATPase motif mutations in Schizosaccharomyces pombe minichromosome maintenance proteins. Genetics 2002; 160:1305 - 18; PMID: 11973289
- Schwacha A, Bell SP. Interactions between two catalytically distinct MCM subgroups are essential for coordinated ATP hydrolysis and DNA replication. Mol Cell 2001; 8:1093 - 104; http://dx.doi.org/10.1016/S1097-2765(01)00389-6; PMID: 11741544
- Bochman ML, Schwacha A. The Mcm complex: unwinding the mechanism of a replicative helicase. Microbiol Mol Biol Rev 2009; 73:652 - 83; http://dx.doi.org/10.1128/MMBR.00019-09; PMID: 19946136
- Treisman R, Ammerer G. The SRF and MCM1 transcription factors. Curr Opin Genet Dev 1992; 2:221 - 6; http://dx.doi.org/10.1016/S0959-437X(05)80277-1; PMID: 1638115
- Koonin EV. A common set of conserved motifs in a vast variety of putative nucleic acid-dependent ATPases including MCM proteins involved in the initiation of eukaryotic DNA replication. Nucleic Acids Res 1993; 21:2541 - 7; http://dx.doi.org/10.1093/nar/21.11.2541; PMID: 8332451
- Iyer LM, Leipe DD, Koonin EV, Aravind L. Evolutionary history and higher order classification of AAA+ ATPases. J Struct Biol 2004; 146:11 - 31; http://dx.doi.org/10.1016/j.jsb.2003.10.010; PMID: 15037234
- Frickey T, Lupas AN. Phylogenetic analysis of AAA proteins. J Struct Biol 2004; 146:2 - 10; http://dx.doi.org/10.1016/j.jsb.2003.11.020; PMID: 15037233
- Erzberger JP, Berger JM. Evolutionary relationships and structural mechanisms of AAA+ proteins. Annu Rev Biophys Biomol Struct 2006; 35:93 - 114; http://dx.doi.org/10.1146/annurev.biophys.35.040405.101933; PMID: 16689629
- Bell SP, Dutta A. DNA replication in eukaryotic cells. Annu Rev Biochem 2002; 71:333 - 74; http://dx.doi.org/10.1146/annurev.biochem.71.110601.135425; PMID: 12045100
- Aparicio OM, Weinstein DM, Bell SP. Components and dynamics of DNA replication complexes in S. cerevisiae: redistribution of MCM proteins and Cdc45p during S phase. Cell 1997; 91:59 - 69; http://dx.doi.org/10.1016/S0092-8674(01)80009-X; PMID: 9335335
- Aparicio OM, Stout AM, Bell SP. Differential assembly of Cdc45p and DNA polymerases at early and late origins of DNA replication. Proc Natl Acad Sci U S A 1999; 96:9130 - 5; http://dx.doi.org/10.1073/pnas.96.16.9130; PMID: 10430907
- Wyrick JJ, Aparicio JG, Chen T, Barnett JD, Jennings EG, Young RA, et al. Genome-wide distribution of ORC and MCM proteins in S. cerevisiae: high-resolution mapping of replication origins. Science 2001; 294:2357 - 60; http://dx.doi.org/10.1126/science.1066101; PMID: 11743203
- Labib K, Tercero JA, Diffley JF. Uninterrupted MCM2-7 function required for DNA replication fork progression. Science 2000; 288:1643 - 7; http://dx.doi.org/10.1126/science.288.5471.1643; PMID: 10834843
- Bowers JL, Randell JC, Chen S, Bell SP. ATP hydrolysis by ORC catalyzes reiterative Mcm2-7 assembly at a defined origin of replication. Mol Cell 2004; 16:967 - 78; http://dx.doi.org/10.1016/j.molcel.2004.11.038; PMID: 15610739
- Randell JC, Bowers JL, Rodríguez HK, Bell SP. Sequential ATP hydrolysis by Cdc6 and ORC directs loading of the Mcm2-7 helicase. Mol Cell 2006; 21:29 - 39; http://dx.doi.org/10.1016/j.molcel.2005.11.023; PMID: 16387651
- Donovan S, Harwood J, Drury LS, Diffley JF. Cdc6p-dependent loading of Mcm proteins onto pre-replicative chromatin in budding yeast. Proc Natl Acad Sci U S A 1997; 94:5611 - 6; http://dx.doi.org/10.1073/pnas.94.11.5611; PMID: 9159120
- Hua XH, Newport J. Identification of a preinitiation step in DNA replication that is independent of origin recognition complex and cdc6, but dependent on cdk2. J Cell Biol 1998; 140:271 - 81; http://dx.doi.org/10.1083/jcb.140.2.271; PMID: 9442103
- Rowles A, Tada S, Blow JJ. Changes in association of the Xenopus origin recognition complex with chromatin on licensing of replication origins. J Cell Sci 1999; 112:2011 - 8; PMID: 10341218
- Blow JJ, Gillespie PJ. Replication licensing and cancer--a fatal entanglement?. Nat Rev Cancer 2008; 8:799 - 806; http://dx.doi.org/10.1038/nrc2500; PMID: 18756287
- Lau KM, Chan QK, Pang JC, Li KK, Yeung WW, Chung NY, et al. Minichromosome maintenance proteins 2, 3 and 7 in medulloblastoma: overexpression and involvement in regulation of cell migration and invasion. Oncogene 2010; 29:5475 - 89; http://dx.doi.org/10.1038/onc.2010.287; PMID: 20661220
- Saydam O, Senol O, Schaaij-Visser TB, Pham TV, Piersma SR, Stemmer-Rachamimov AO, et al. Comparative protein profiling reveals minichromosome maintenance (MCM) proteins as novel potential tumor markers for meningiomas. J Proteome Res 2010; 9:485 - 94; http://dx.doi.org/10.1021/pr900834h; PMID: 19877719
- Remus D, Diffley JF. Eukaryotic DNA replication control: lock and load, then fire. Curr Opin Cell Biol 2009; 21:771 - 7; http://dx.doi.org/10.1016/j.ceb.2009.08.002; PMID: 19767190
- Gregan J, Lindner K, Brimage L, Franklin R, Namdar M, Hart EA, et al. Fission yeast Cdc23/Mcm10 functions after pre-replicative complex formation to promote Cdc45 chromatin binding. Mol Biol Cell 2003; 14:3876 - 87; http://dx.doi.org/10.1091/mbc.E03-02-0090; PMID: 12972571
- Tuteja N, Tran NQ, Dang HQ, Tuteja R. Plant MCM proteins: role in DNA replication and beyond. Plant Mol Biol 2011; 77:537 - 45; http://dx.doi.org/10.1007/s11103-011-9836-3; PMID: 22038093
- Dang HQ, Tran NQ, Gill SS, Tuteja R, Tuteja N. A single subunit MCM6 from pea promotes salinity stress tolerance without affecting yield. Plant Mol Biol 2011; 76:19 - 34; http://dx.doi.org/10.1007/s11103-011-9758-0; PMID: 21365356
- Arnot DE, Gull K. The Plasmodium cell-cycle: facts and questions. Ann Trop Med Parasitol 1998; 92:361 - 5; http://dx.doi.org/10.1080/00034989859357; PMID: 9683889
- Tuteja R. Malaria - an overview. FEBS J 2007; 274:4670 - 9; http://dx.doi.org/10.1111/j.1742-4658.2007.05997.x; PMID: 17824953
- Bannister L, Mitchell G. The ins, outs and roundabouts of malaria. Trends Parasitol 2003; 19:209 - 13; http://dx.doi.org/10.1016/S1471-4922(03)00086-2; PMID: 12763426
- Li JL, Cox LS. Identification of an MCM4 homologue expressed specifically in the sexual stage of Plasmodium falciparum.. Int J Parasitol 2001; 31:1246 - 52; http://dx.doi.org/10.1016/S0020-7519(01)00237-5; PMID: 11513894
- Patterson S, Robert C, Whittle C, Chakrabarti R, Doerig C, Chakrabarti D. Pre-replication complex organization in the atypical DNA replication cycle of Plasmodium falciparum: characterization of the mini-chromosome maintenance (MCM) complex formation. Mol Biochem Parasitol 2006; 145:50 - 9; http://dx.doi.org/10.1016/j.molbiopara.2005.09.006; PMID: 16257456
- Ramakrishnan C, Dani VS, Ramasarma T. A conformational analysis of Walker motif A [GXXXXGKT (S)] in nucleotide-binding and other proteins. Protein Eng 2002; 15:783 - 98; http://dx.doi.org/10.1093/protein/15.10.783; PMID: 12468712
- Komamura-Kohno Y, Tanaka R, Omori A, Kohno T, Ishimi Y. Biochemical characterization of fragmented human MCM2. FEBS J 2008; 275:727 - 38; http://dx.doi.org/10.1111/j.1742-4658.2007.06239.x; PMID: 18190532
- Lin DI, Aggarwal P, Diehl JA. Phosphorylation of MCM3 on Ser-112 regulates its incorporation into the MCM2-7 complex. Proc Natl Acad Sci U S A 2008; 105:8079 - 84; http://dx.doi.org/10.1073/pnas.0800077105; PMID: 18524952
- Li J, Deng M, Wei Q, Liu T, Tong X, Ye X. Phosphorylation of MCM3 protein by cyclin E/cyclin-dependent kinase 2 (Cdk2) regulates its function in cell cycle. J Biol Chem 2011; 286:39776 - 85; http://dx.doi.org/10.1074/jbc.M111.226464; PMID: 21965652
- Komamura-Kohno Y, Karasawa-Shimizu K, Saitoh T, Sato M, Hanaoka F, Tanaka S, et al. Site-specific phosphorylation of MCM4 during the cell cycle in mammalian cells. FEBS J 2006; 273:1224 - 39; http://dx.doi.org/10.1111/j.1742-4658.2006.05146.x; PMID: 16519687
- Hughes CR, Guasti L, Meimaridou E, Chuang CH, Schimenti JC, King PJ, et al. MCM4 mutation causes adrenal failure, short stature, and natural killer cell deficiency in humans. J Clin Invest 2012; 122:814 - 20; http://dx.doi.org/10.1172/JCI60224; PMID: 22354170
- Gineau L, Cognet C, Kara N, Lach FP, Dunne J, Veturi U, et al. Partial MCM4 deficiency in patients with growth retardation, adrenal insufficiency, and natural killer cell deficiency. J Clin Invest 2012; 122:821 - 32; http://dx.doi.org/10.1172/JCI61014; PMID: 22354167
- Ferguson RL, Maller JL. Cyclin E-dependent localization of MCM5 regulates centrosome duplication. J Cell Sci 2008; 121:3224 - 32; http://dx.doi.org/10.1242/jcs.034702; PMID: 18799789
- Ferguson RL, Pascreau G, Maller JL. The cyclin A centrosomal localization sequence recruits MCM5 and Orc1 to regulate centrosome reduplication. J Cell Sci 2010; 123:2743 - 9; http://dx.doi.org/10.1242/jcs.073098; PMID: 20663915
- Giaginis C, Giagini A, Tsourouflis G, Gatzidou E, Agapitos E, Kouraklis G, et al. MCM-2 and MCM-5 expression in gastric adenocarcinoma: clinical significance and comparison with Ki-67 proliferative marker. Dig Dis Sci 2011; 56:777 - 85; http://dx.doi.org/10.1007/s10620-010-1348-5; PMID: 20694513
- Zhang J, Yu L, Wu X, Zou L, Sou KK, Wei Z, et al. The interacting domains of hCdt1 and hMcm6 involved in the chromatin loading of the MCM complex in human cells. Cell Cycle 2010; 9:4848 - 57; http://dx.doi.org/10.4161/cc.9.24.14136; PMID: 21099365
- Xu J, Zhang S, You C, Huang S, Cai B, Wang X. Expression of human MCM6 and DNA Topo II alpha in craniopharyngiomas and its correlation with recurrence of the tumor. J Neurooncol 2007; 83:183 - 9; http://dx.doi.org/10.1007/s11060-006-9284-0; PMID: 17410335
- Toyokawa G, Masuda K, Daigo Y, Cho HS, Yoshimatsu M, Takawa M, et al. Minichromosome Maintenance Protein 7 is a potential therapeutic target in human cancer and a novel prognostic marker of non-small cell lung cancer. Mol Cancer 2011; 10:65; http://dx.doi.org/10.1186/1476-4598-10-65; PMID: 21619671
- Doerig C, Chakrabarti D, Kappes B, Matthews K. The cell cycle in protozoan parasites. Prog Cell Cycle Res 2000; 4:163 - 83; http://dx.doi.org/10.1007/978-1-4615-4253-7_15; PMID: 10740824
- Gozuacik D, Chami M, Lagorce D, Faivre J, Murakami Y, Poch O, et al. Identification and functional characterization of a new member of the human Mcm protein family: hMcm8. Nucleic Acids Res 2003; 31:570 - 9; http://dx.doi.org/10.1093/nar/gkg136; PMID: 12527764
- Volkening M, Hoffmann I. Involvement of human MCM8 in prereplication complex assembly by recruiting hcdc6 to chromatin. Mol Cell Biol 2005; 25:1560 - 8; http://dx.doi.org/10.1128/MCB.25.4.1560-1568.2005; PMID: 15684404
- Kinoshita Y, Johnson EM, Gordon RE, Negri-Bell H, Evans MT, Coolbaugh J, et al. Colocalization of MCM8 and MCM7 with proteins involved in distinct aspects of DNA replication. Microsc Res Tech 2008; 71:288 - 97; http://dx.doi.org/10.1002/jemt.20553; PMID: 18072282
- Yoshida K. Identification of a novel cell-cycle-induced MCM family protein MCM9. Biochem Biophys Res Commun 2005; 331:669 - 74; http://dx.doi.org/10.1016/j.bbrc.2005.03.222; PMID: 15850810
- Lutzmann M, Maiorano D, Méchali M. Identification of full genes and proteins of MCM9, a novel, vertebrate-specific member of the MCM2-8 protein family. Gene 2005; 362:51 - 6; http://dx.doi.org/10.1016/j.gene.2005.07.031; PMID: 16226853
- Lutzmann M, Méchali M. MCM9 binds Cdt1 and is required for the assembly of prereplication complexes. Mol Cell 2008; 31:190 - 200; http://dx.doi.org/10.1016/j.molcel.2008.07.001; PMID: 18657502
- Fletcher RJ, Bishop BE, Leon RP, Sclafani RA, Ogata CM, Chen XS. The structure and function of MCM from archaeal M. Thermoautotrophicum.. Nat Struct Biol 2003; 10:160 - 7; http://dx.doi.org/10.1038/nsb893; PMID: 12548282
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem 2004; 25:1605 - 12; http://dx.doi.org/10.1002/jcc.20084; PMID: 15264254
- Pereverzeva I, Whitmire E, Khan B, Coué M. Distinct phosphoisoforms of the Xenopus Mcm4 protein regulate the function of the Mcm complex. Mol Cell Biol 2000; 20:3667 - 76; http://dx.doi.org/10.1128/MCB.20.10.3667-3676.2000; PMID: 10779356
- Ishimi Y, Komamura-Kohno Y. Phosphorylation of Mcm4 at specific sites by cyclin-dependent kinase leads to loss of Mcm4,6,7 helicase activity. J Biol Chem 2001; 276:34428 - 33; http://dx.doi.org/10.1074/jbc.M104480200; PMID: 11454864
- Blom N, Sicheritz-Pontén T, Gupta R, Gammeltoft S, Brunak S. Prediction of post-translational glycosylation and phosphorylation of proteins from the amino acid sequence. Proteomics 2004; 4:1633 - 49; http://dx.doi.org/10.1002/pmic.200300771; PMID: 15174133
- Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, et al. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res 2009; 37:Web Server issue W202-8; http://dx.doi.org/10.1093/nar/gkp335; PMID: 19458158