Abstract
There have been a wide variety of approaches for handling the pieces of DNA as the “unplugged” tools for digital information storage and processing, including a series of studies applied to the security-related area, such as DNA-based digital barcodes, water marks and cryptography. In the present article, novel designs of artificial genes as the media for storing the digitally compressed data for images are proposed for bio-computing purpose while natural genes principally encode for proteins. Furthermore, the proposed system allows cryptographical application of DNA through biochemically editable designs with capacity for steganographical numeric data embedment. As a model case of image-coding DNA technique application, numerically and biochemically combined protocols are employed for ciphering the given “passwords” and/or secret numbers using DNA sequences. The “passwords” of interest were decomposed into single letters and translated into the font image coded on the separate DNA chains with both the coding regions in which the images are encoded based on the novel run-length encoding rule, and the non-coding regions designed for biochemical editing and the remodeling processes revealing the hidden orientation of letters composing the original “passwords.” The latter processes require the molecular biological tools for digestion and ligation of the fragmented DNA molecules targeting at the polymerase chain reaction-engineered termini of the chains. Lastly, additional protocols for steganographical overwriting of the numeric data of interests over the image-coding DNA are also discussed.
Introduction
Natural genes principally encode for proteins.Citation1 In contrast, the aim of the present perspective article is to propose a novel artificial gene model for bio-computing purpose, by enabling the storage of digitally compressed imaging data in the edittable DNA molecules. Furthermore, the proposed system allows cryptographical application of DNA. As a model case of image-coding DNA technique application, numerically and biochemically combined protocols are employed for ciphering the given “passwords” and/or numeric data using DNA sequences.
For ciphering the message or “passwords” of importance which should not be kept as written information on papers or on the computers (and related digitalized memories) for security reasons, alternative means of coding, editing, memorizing (preserving), coping and decoding of information must be considered. Use of bio-macromolecules such as DNA is one of likely choices for cryptographically encoding the information of interest.
DNA is a nucleic acid which was first isolated one and half centuries ago.Citation1 Identification of DNA as the carrier of genetic information was first reported in 1944.Citation2 In 1953, James D. Watson and Francis Crick suggested the double-helix model of DNA structure for the first time.Citation3 Since then, the age of molecular biology has drastically opened. It is now widely accepted that DNA contains the genetic information instructing the manners to be used in the developments and functioning in all living cellular organisms and DNA virus. Within the cells of living organisms, amazingly long chains of DNA are packed into the compact structures called chromosomes.Citation4 The size of information coded within the single set of molecules inside each of the micrometer-sized human cells exceeds ca. 3-billion base pairs of DNA,Citation5 thus large scale integration has been naturally manifested in the course of evolution. Therefore, it is tempting to speculate that DNA can be used as the means of “unplugged” information storage and processing.
There have been a wide variety of approaches for handling the pieces of DNA as the “unplugged” tools for digital information storage and processing. Rauhe et al.Citation6 has reported their attempt for creating digital DNA molecules representing the binary data structures based on the programmable self-assembly nature of DNA oligo-nucleotides. In their work, plasmid (circular double stranded DNA) was used as the “memory” with programmability, which allowed isolation, amplification and reading out based on the common genetic techniques.
However, in their approach, oligo-nucleotide sequences rather than single bases such as adenine (A), cytosine (C), guanine (G) and thymine (T) were used as bits, thus the size of information to be encoded or handled must be largely limited. Obviously, handling of each nucleotide base as a bit is desirable for developing the novel encoding system applicable to large-sized data encoding and processing. Thus, the main purpose of the present article is to propose a novel upscalable data-encoding rule for DNA-based imaging. Although the examples chosen are not very practical or realistic, the handling of simple and minimal sized information to be coded in DNA may best explain the systems proposed.
In addition to the size of information storage capacity, DNA-based informatics attacted the researchers from the points of computability and security of data. Recently, a series of approaches employing the DNA sequences as novel informative codes or security tools were reported. Such studies include the use of DNA molecules as digital barcodes,Citation6 digital water marks,Citation7 and the media for liquid computing and cryptography.Citation8,Citation9 In fact, the second purpose of the present article is to describe an example of DNA-based semi-numerical and semi-biochemical cryptography combining the above newly proposed DNA encoding/decoding rules and molecular biological techniques.
Our protocols presented here enables the ciphering and decoding of the original “password” through both numerical and biochemical manners, by encoding each letter from the given “passwords” as the font images on the separate DNA molecules based on the newly proposed run-length encoding (RLE) system. The RLE is the numerical part of cryptographical approach since the original images could not be obtained without knowing the algorithm employed. Furthermore, even after the images of letter fonts were successfully decoded, they are merely the pieces of letters but not the original “passwords” of interest. The non-coding regions on the DNA molecules are designed for biochemical editing and remodeling processes through which the unveiling of the hidden order of letters to be aligned as the original “passwords” can be achieved. These processes require the molecular biological tools for digestion and ligation of the fragmented DNA at the polymerase chain reaction (PCR)-engineered termini of the chains.
As above, in addition to the expected decoding procedure (reading of the images from the binary codes written on the DNA), biochemical data conjunction process for properly lining the letters to form the coded words or sentences are necessarily required. Furthermore, additional protocols for steganographically overlaying the numeric data (decimal numbers) of interests within the image-coding DNA are also proposed and discussed.
Proposed Structures for Artificial Genes Coding for the Images
The segments of DNA found in nature conveying the genetic information are called genes. For regulated function of the genes, the presence of non-coding regions of DNA with specific structures is required (). While the genes in the biological system principally code for the proteins (via transcription from DNA to RNA and translation from RNA to amino acid sequence), the artificial genes designed here encode for the compressed data for images (such as font images for examples). Another key difference between the natural genes and artificial image-coding genes are the modes of data encoding. While the former uses the strings of nucleotide bases, the latter system employs RLE rule as discussed later.
Figure 1. The proposed structures of the DNA fragments coding for the images. (A) Chemical structure of DNA (only single stranded form is shown). (B) Schematic model for the structure of a gene and regulatory sequences coded on DNA. (C) Schematic model for the structure of an “image-coding DNA unit” newly proposed here. The numbers, 5′ and 3′ at the ends of DNA indicate the orientation of the chains (coding and reading start from 5′-terminal toward the 3′-terminal). Black box at the center of the DNA chain stands for the image coding region. Within the white box corresponding to the non-coding regions, five different elements are embedded namely: (1) the positional markers; (2) the tags for DNA-based “password” editing procedures such as cutting and pasting; (3) the starting points and end points for the copying events; (4) the labeling required for filing (or addressing); and (5) embedment of hidden information (e.g., steganography).
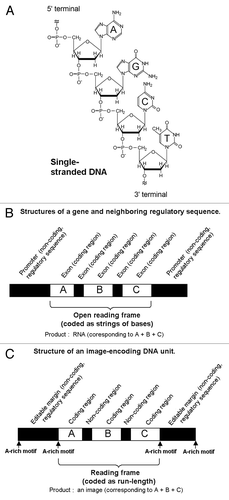
By analogy to the simplified gene structure, the image coding regions analogous to the genes within the synthetic DNA sequence were conjugated with and between the non-coding DNA sequences (). The non-coding DNA sequences can be dissected into the sequence found within the reading frame and those outside the frame. Former resembles the “introns” which are fragmented DNA sequences inserted among the coding sequences called “exons” in natural genes; and the latter resembles the regulatory sequence motifs found in the promoter regions outside the natural genes. Above non-coding DNA sequences designed for the image coding/editing purposes play important roles such as: (1) the positional markers; (2) the tags for editing procedures such as cutting and pasting; (3) the starting points and end points for the sequence copying events by PCR; (4) the labeling required for filing or addressing, and as discussed later; for (5) embedment of hidden information (e.g., steganography).
In case of natural genes, fragmented information coded on DNA are copied (transcripted) into a single strand of mRNA (mRNA), on which the coded information is still in the fragmented forms. For obtaining the correct products (proteins) of the genetic codes from the fragmented information, mRNAs are subjected to further process (splicing) for removal of non-coding sequences prior to the translation events on ribosomes.
In contrast, the fragmented information embedded on the “Image-coding” DNA can be integrated simply after decoding of the information. Therefore, there is no need for splicing or molecular process directly jointing the coded regions on a single DNA strand. However, when required, actual cut-and-paste processing for substancially editing the coded sequence could be performed based on the desigin of the original DNA units as discussed below.
Model Demonstration with Font Images
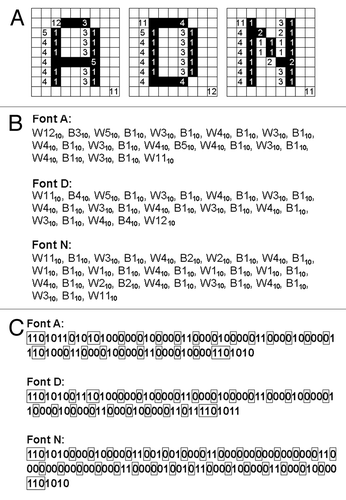
Assuming that the word ADN (standing for Aircraft Data Network; http://www.arinc.com) is the model “password” to be ciphered in DNA (within the image coding regions); these letters (A, D and N) could be separately converted as the fragments of font images (). In the present work, the DNA sequence coding for each of these font images is refered to as an artificial gene. By reading from the image pixel at the left corner on the top of the 9 × 9 pixel squares for A, D and N; the font image can be translated to the string of binary numbers 1 and 0 (1 for black and 0 for white) as shown in .
Run-Length Encoding (RLE) System
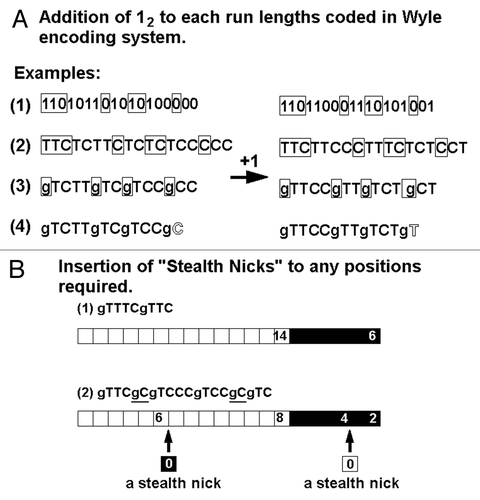
The nucleotide sequences corresponding to the image codes should be placed within the coding regions on artificial genes. In order to design the image coding region, the encoding system to be employed must be determined first. Obviously, the simplest way for digitalization may be direct translation of the image of interest to be encoded by DNA, into the strings of binary numbers (12 and 02). However, for handling of large-sized digitalized data, RLE procedures are highly beneficial for saving the number of nucleotides required, by effectively compressing the data. Among RLE systems, a classical system proposed by Wyle et al.Citation10 has been widely used for long period, especially for encoding the image for facsimiles.
Let’s start developing the novel RLE system for DNA-based imaging by considering the basic idea from the Wyle system. In , the strings of numbers coding for the font images “A,” “D,” and “N” were converted to a series of run-lengths based on the Wyle encoding system. The run-lengths found in the font images in can be summarized as a series of white and black runs such as “white × 13 (W13) + black × 3 (B3) +…” and so on (). Assuming that the first run can be always a “white run” and alternate chains of black and white runs continues, above description can be simplified as the sequence of run-length numbers. For expressing such a series of run-lengths using binary numbers, the Wyle encoding system employs the pairs of a prefix (defining the digit numners required for the run) and a run-length code (). Note that there are some variations in the commonly employed Wyle encoding systems, e.g., 510 can be expressed as (10, prefix) + (100, run-length) or (10, prefix) + (00, run-length). In the former example, prifix “10” defines that the run should be a 3-digit binary number such as 100. This article employs the former encoding system as a starting point.
Figure 3. The model “password” encoded by a RLE system (Wyle encoding system). (A) The run-lengths consisting the fonts of model letters A (left), D (middle) and N defined on the 9 × 9 block-square were counted. (B) The blocks of black and white colors were converted to run-lengths. The numbers shown are in decimal. (C) The run-lengths forming the letter fonts were expressed with Wyle encoding system. Boxed numbers, prefixes, other number, run-length. The numbers shown are binary.
Proposed RLE Rules for Image Encoding DNA
In order to develop the DNA-based image coding system, RLE must be expressed using the DNA bases. As the very first step, the sequences of binary numbers encoding for the font images based on Wyle encoding system were simply converted to the sequence of DNA bases (). Here, both the prefixes and run-length codes were expressed with thymine (T) and cytosine (C), in place of 1 and 0, respectively (other bases could be chosen of course).
Figure 4. Wyle encoding system-based Run-length encoding approaches using DNA. (A) Prototype code 0.1. Note that 1 and 0 in wyle encoding system are simply converted to T and C. Prefixes are boxed. (B) Prototype code 0.2. Again, prefixes are boxed. The prefixes preceding the run-length codes are expressed with single guanine base, G. (C) Prototype code 0.3. Outlined letters indicate the positions shorten by new coding rule.
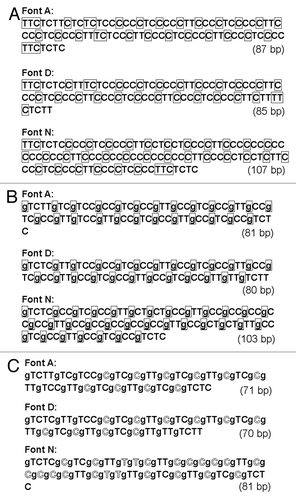
In case of coding for long run-lengths, the prefixes required tend to be quite long one. For examples, the prefixes preceding the 3-, 4-, 5-, 6-, 7-, or 8-digit binary numbers (for expressing the run-lengths ranged from 510 to 25610) would be TC, TTC, TTTC, TTTTC, TTTTTC, or TTTTTTC (10, 110, 1110, 11110, 111110, or 1111110), respectively. In fact, a prefix merely functions as the gap separating two different run-length codes, thus, this can be replaced with a single base, since DNA has two additional sets of bases such as guanine (G). In , all the prefixes in the RLE were replaced with “g” (gaps by G). Note that the above modified Wyle encoding system still uses the expression with “prefix + CC (00)” and “prefix + CT (01)” for coding for short runs such as single-pixelled and two-pixelled runs, respectively. In fact, in the revised system, the initial C (0) in each of run-length codes is no longer required ().
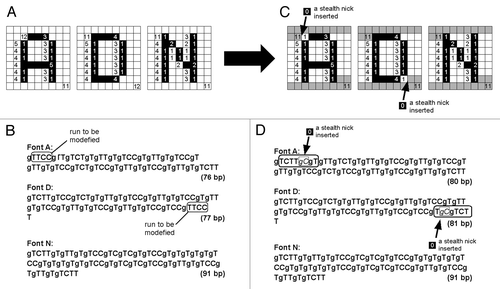
As most of readers are aware of the fact that both Wyle encoding system and the derived temporal system for DNA described above display the numbers (n) as (n-1), thus instead of 110, 210, 310, 410 and so on, C (02), T (12), TC (102), TT (112) and so on, respectively are used. Therefore, these systems prohibit the use of zero as 0 (or C). For allowing the novel code for length-less runs, the newly revised system expresses the run-length as it is, thus (n), rather than (n-1), as shown in . As a consequence, now an insertion of the code (gC) encoding for zero (length-less run) into the DNA sequence is practically allowed. By inserting such codes for length-less runs into any sites of interest, new positional markers can be created without interrupting the apparent run-lengths displayed in the decoded images (). The interruption of the coded run-lengths by insertion of single gC or odd numbered gCs could not be graphically detected after decoding of the images. Therefore such interruptions are now referred to as “stealth nicks.” Effective uses of such stealth nicks will be discussed in the later sections.
Allocation of A-rich Motifs in the Non-Coding Margins
Since the RLE system proposed here employs only C and T as the binary (02, 12) and G, as for the gaps (g) among four members of DNA bases, even single A is not used in the coding regions. Therefore, the use of A-rich motifs (referred to as boxes) can be the markers distinguishing the coding regions and the regulatory marginal regions within the coding strand of DNA ().
Many examples of A-rich motifs (boxes) found in the promoter regions of the natural genes, such as TATA box, can be the model for the motifs used in our system. As the margins can be the scaffold for molecular amplification by PCR, certain ratios of C and G over A and T must be designed for better annealing with the primers. Thus, in the present model, GAGAGA and AGAGAG boxes were allocated as the markers for the starting and end points of the coding regions, respectively. Furthermore, within the marginal regions defined by these GA-rich motifs, the tags for editing procedures such as cutting and pasting, scaffold for PCR and the labeling required for filing or addressing can be installed when required.
Taylor-Made PCR Reactions for Introducing the Restriction Enzyme Recognition Sites
Bacterial world employs a series of enzymes called restriction enzymes, for cutting out (restrict) the pathogenic DNAs (mostly double-stranded DNAs) at or near the specific site of recognition. These enzymes are now used as common tools for genetic engineering. The resultant pieces of DNA released after enzymatic digestion can be readily rejointed (ligated) when these DNA fragments possess terminal overhangs complementary to each other. RothemundCitation11 suggested that these operations can be applied to the DNA-based data processing, especially DNA-based computing.
Among the restriction enzymes, some combinations of enzymes can be used for the digestion of multiple restriction sites at once in the same reaction buffer; for examples restriction enzymes SacI and KpnI can be jointly used in the low salt buffer, EcoR I, Hind III and TaqI can be jointly used in the medium salt buffer, and BamHI, ClaI and PstI can be jointly used in the high salt buffer. Therefore, combinations of enzymes which can be used in the identical conditions are highly recommended for simplified preparation for the orientation-designed DNA ligation.
As shown in , any desired restriction enzyme recognition sites can be introduced in the non-coding marginal regions of the image-coding DNA molecules, by designing the pairs of PCR primers (oligo DNA) containing both the restriction site sequences (5 to 6 bases) and the oligo-DNA sequence complementary to the marginal regions oligo-DNA sequence. Therefore, the combinations of the PCR primers used would be the necessary “key” for obtaining the solution to the biochemical decoding operations ().
Figure 6. Taylor-made PCR for introducing the restriction sites of interest for designed DNA-chain conjugation. Note that the presence of complementary chains of DNA is not shown on the illustrations for simplification. (A) Separately coded letter images and a list of uncountable choices (likely passwords). (B) Preparation of PCR primers for designed DNA digestion. (C) Restriction enzyme recognizition sites PCR-dependently created at terminals of DNA. (D) Digestion by selected restriction enzymes. (E) Formation of novel molecule during the “password” decoding process. M, E and H strand for marginal regions, EcoR I recognition sites and Hind III recognitions sites, respectively.
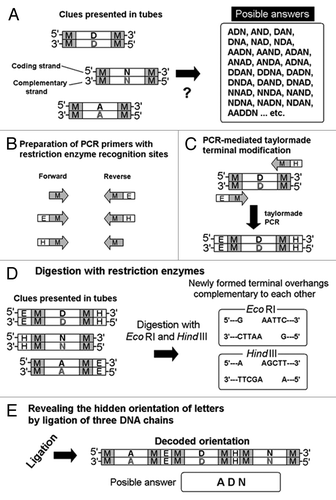
Protection of the Coding region from the Digestion by Restriction Enzymes
As discussed above, DNA can be selectively and specifically digested using the restriction enzymes which recognize the specific sites on the DNA. Due to the coding rule design, the coding regions lack A. This feature can be used for selected digestion of DNA only within the marginal regions avoiding the unexpected cut in the coding regions. However, the absence of A in the coding region does not mean that coding sequences are safe against A-recognizing enzymes since A complementary to T appears in the complementary chains even within the coding regions. Therefore, a series of enzymes which recognize the sequences containing both A and T at the same time on the same chains were chosen in the model demonstration shown in . The recognition sites for EcoR I and Hind III were introduced into the DNA chains by PCR. EcoR I recognizes 5′-GAATTC-3′ on one chain of double stranded DNA and at the same time 3′-CTTAAG-5′ on the complementary chain is recognized, to cut the phosphate links between G and A, thus releasing two double stranded DNA with terminal overhangs complementary to each other (, right). Digestion with Hind III also results in generation of such DNA chains with “adhesive ends.”
Ligation Reveals the Hidden Orientation of the Letters
Following digestion of PCR-engineered DNA chains with restriction enzymes, resultant DNA chains with “adhesive ends” must be subjected to the ligation by ligase, to reveal the original orientation of the letters within the “password” (). After obtaining the expected size of DNA chain (examined on agarose gel), isolated band should be used for DNA sequencing. As illustrated in , there were uncountable numbers of candidate words (such as D-N-A, A-N-D, N-D-A, etc.) but after application of the molecular genetics tools, the “password” was determined to be A-D-N (). In this case, sets of primers and/or sets of restriction enzymes matching the molecular design would be the actual keys to the answer.
Use of “Stealth Nicks”—Case 1: Adjustment of Run-Length at Termini of Coding Regions
As the marginal regions share common motifs of oligo-nucleotide sequences to be used as the starting point for PCR, the role for the marginal regions as the scaffolds for PCR was described above. The oligo-nucleotide sequences at two termini of the font image-coding region in each DNA molecule can be additionally used as the common scaffold for PCR, after certain modifications of the length of the first and last runs for RLE were made. As shown in , the first and last runs of RLE in three of font image-coding DNA molecules were adjusted be identical in length (i.e., 11), by inserting the single stealth nicks (gC) encoding for length-less runs into the initial run of the font A-coding DNA and the last run of the font D-coding DNA. In , the first and last runs after the length adjustment with stealth nick insertions are highlighted as 11 consecutive dark pixels. Now the initial and the last runs can be commonly coded as gTCTT. This allows designing common primers for PCR, for examples, the forward and reverse primers for all font images now commonly contain gTCTTg and gTCTT, respectively.
Figure 7. Designing the common terminal structures for all of the letter-coding DNA chains by insertions of the pair of G (guanine used as a gap) and C (cytosine) as a “stealth nick” within the font-coding reading frames. After insertions of gC to Font A and Font D, at 5′-termini and at 3′-termini, the sequences gTCTTg and gTCTT, respectively, are common to all DNA chains.
Use of “Stealth Nicks”—Case 2: Steganographic Numerical Data Overwriting
Although people have hidden secrets in plain sight throughout the ages, the recent growth in computational power and technology has propelled it to the forefront of today's security techniques.Citation12 These approaches are now called steganography, as the key concepts are illustrated in . Here, additional protocols for overwriting of the numeric data of interests over the font image-coding DNA molecules based on newly proposed steganographic approaches using the stealth nick-inserted RLE are discussed in this section.
Figure 8. DNA-encoded image-based steganography. (A) Generalized concept for steganography overlying the numbers of interest under the media (cover media). (B) DNA RLE-based steganographic approach.
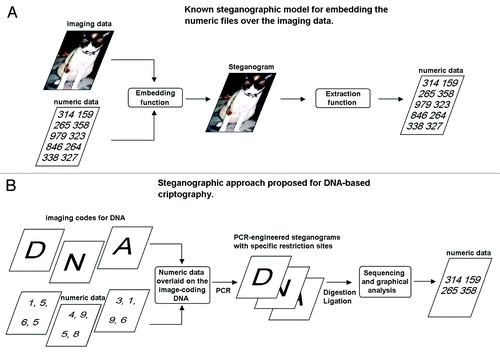
Steganography is the art and science of hiding communication; a steganographic system thus embeds hidden content in unremarkable cover media.Citation12 Wong et al.Citation13 first developed a steganographic algorithm based on DNA, which is able to store data in living organisms from which data can be extracted by PCR. In this case, the information of the interest is hidden as the plasmid-encoded sequence, thus two set of information, namely: (1) original genome; and (2) newly installed circular DNA are coexisting in the cells. In contrast, the approach present here tries hiding numeric information over image coding information within the same molecules of DNA ().
Usually, the information-hiding process in a steganographic system starts by identifying a cover medium's redundant bits (those that can be modified without destroying that medium's integrity) and the embedding process creates a stego medium by replacing these redundant bits with data for the hidden message.Citation12
demonstrates the steganographic approach applicable in the image-coding DNA model, by inserting some length-less runs (stealth nicks) into any positions on the 9 × 9 imaginary square coded by RLE. As discussed earlier (), by inserting the stealth nicks into the RLE data, the run-lengths can be interrupted without distorting the encoded image. The number of stealth nicks inserted at one position can be multiple but must be odd numbered (gCn, n = 1, 3, 5, 7…). By marking the horizontal lines of interest on the 9 × 9 square (showing the font-images), decimal numbers from 110 to 910 can be encoded. By this way, 010 can be coded as the absence of the stealth nick on any line. The hierarchy can be brought on among the numbers embedded as the size of nick repeat, with the single nick as the highest and the septuple nicks as the lowest. Since the three independent font images (coded on separate, molecules of DNA) can be joined as single chain of double stranded DNA, thus stego-medium for the hidden numeric codes can be also saved separately and decoded by molecular ligation. Taken together, the sequence of the ciphered numbers can be determined according to: (1) the restriction enzyme-dependently determined order (A→D→N) of DNA chains conjugated by the ligation; and (2) the hierarchy of the nicks inserted, as the procedures are summarized in . Therefore, the 12-digit number steganographically hidden in the DNA sequence can be extracted and determined to be 3141 5926 5358.
Figure 9. Steganographic insertion of stealth nicks (gC)s into the font image-coding DNA sequences used as cover media (A, D and N), at the positions corresponding to numeric codes. (A) Insertion of stealth nicks into the font images coded by DNA. Coded image even after insertion of stealth nicks (left). Images visualizing the position of stealth nick insertions (right). (B) Steganographically modified DNA sequences coding for the font images (A, D and N) with RLE. Italicized letters indicate the insertion of stealth nicks into the original DNA sequences.

Table 1. Hidden numbers steganographically encoded by the positions of stealth nick inserted into the DNA RLEs coding for the font images
Discussion
In the present article: (1) RLE data coding rules for DNA-based informatics; (2) molecular biological ciphering techniques; and (3) steganographical protocols handling numbers under within the image-coding DNA were proposed. Among the three topics covered here, the author would like to emphasize the proposal of new RLE rules for DNA informatics, thus large space was dedicated to this topic.
The image-coding rules designed for encoding and storage of the image data using DNA are summarized in . In biological system, the chains of natural gene-coding DNA are merely the strings of 4 digit data (A, T, G, C) naturally designed for encoding the sequential information to be scripted as the strings of RNA, and literally translated into the strings of amino acids to form the proteins. By learming from the nature, it is natural to use the DNA for encoding the strings of digits, and thus, many researchers and engineers suggested that DNA can be the media for coding such strings of data.Citation4
Table 2. Comparison of data compressing RLE protocols for DNA-coded images
In the present study, the author showed a series of proposals for handling the data to be coded on DNA, not as the string of bits but the run-lengths enabling the compression of data with a minimal number of bases within DNA. This allows the encoding of the images at the size readily engineered without technical difficulty. In case of RLE, the original data before compression can be hardly decodable unless algorithm is open to those seeking for it. Therefore, employment of RLE can be considered as the numerical part of cryptography presented here.
In addition, the decoded images simply show some fragments of characters or letters at this stage. Therefore, unless additional editing and processing by PCR-dependent modification, enzymatic digestion and ligation of DNA at the imbedded tag sequences, the orientation of the letters or steganographically hidden numbers cannot be decodable. Furthermore, security of the image can be improved by inserting the length-less runs within the image coded by RLE as steganographic information.
Perspectives
At the interface of biological science and informatics, applications of biological molecules such as nucleotides and proteins or even living organisms: toward (1) the micro- and plant-biorobotics based on automata theory;Citation14 (2) arithmetic and natural computing models;Citation15 and (3) “unplugged” data storage have been recently attempted.
In the present article, novel image-coding RLE rule combined with conventional molecular biological tools are proposed. The most notable aspect found in the proposed techniques is the use of “length-less runs” referred to as stealth nicks which could be inserted into the cover media as steganographic tags. In addition, the stealth nicks can be applicable for PCR-mediated editing of DNA sequence by using the stealth nicks as the tag for primer designs. Furthermore, the stealth nick-based tagging technique may contribute not only to the DNA-based informatics but also to the newly developed DNA-based bioengineering for detection of low concentration of chemicals in the aquatic environments.Citation16,Citation17
| Abbreviations: | ||
| DNA | = | deoxyribonucleic acid |
| PCR | = | polymerase chain reaction |
| RLE | = | run-length encoding |
Acknowledgments
This work was supported by a grant of Regional Innovation Cluster program and a Grants-in-Aid for Scientific Research by Ministry of Education, Culture, Sports, Science and Technology (MEXT), Japan (Research Project Number:23656495).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Dahm R. Discovering DNA: Friedrich Miescher and the early years of nucleic acid research. Hum Genet 2008; 122:565 - 81; http://dx.doi.org/10.1007/s00439-007-0433-0; PMID: 17901982
- Avery OT, Macleod CM, McCarty M. Studies on the chemical nature of the substance inducing transformation of pneumococcal types. Inductions of transformation by a deoxyribonucleic acid fraction isolated from pneumococcus type III. J Exp Med 1944; 79:137 - 58; http://dx.doi.org/10.1084/jem.79.2.137; PMID: 19871359
- Watson JD, Crick FHC. Molecular structure of nucleic acids; a structure for deoxyribose nucleic acid. Nature 1953; 171:737 - 8; http://dx.doi.org/10.1038/171737a0; PMID: 13054692
- Carlson EA. Defining the gene: an evolving concept. Am J Hum Genet 1991; 49:475 - 87; PMID: 1867208
- Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, et al. The sequence of the human genome. Science 2001; 291:1304 - 51; http://dx.doi.org/10.1126/science.1058040; PMID: 11181995
- Rauhe H, Vopper G, Feldkamp U, Banzhaf W, Howard JC. Digital DNA molecules. Proc. 6th DIMACS Workshop on DNA Based Computers; Leiden, Netherlands, 2000; 13-17.
- Heider D, Barnekow A. DNA-based watermarks using the DNA-Crypt algorithm. BMC Bioinformatics 2007; 8:176; http://dx.doi.org/10.1186/1471-2105-8-176; PMID: 17535434
- Clelland CT, Risca V, Bancroft C. Hiding messages in DNA microdots. Nature 1999; 399:533 - 4; http://dx.doi.org/10.1038/21092; PMID: 10376592
- Gehani A, LaBean TH, Reif JH. DNA-based cryptography. Discr Math Theor Comput Sci 2000; 54:233 - 49
- Wyle H, Erb T, Banow R. Reduced-time facsimile transmission by digital coding. IRE Trans Commun Syst 1961; 9:215 - 22; http://dx.doi.org/10.1109/TCOM.1961.1097692
- Rothemund PWKA. DNA and restriction enzyme implimentation of Turing machines. Discr Math Theor Comput Sci 1996; 27:75 - 119
- Provos N, Honeyman P. Hide and seek: an introduction to steganography. Secur Priv IEEE 2003; 1:32 - 44; http://dx.doi.org/10.1109/MSECP.2003.1203220
- Wong PC, Wong KK, Foote H. Organic data memory using the DNA approach. Commun ACM 2003; 46:95 - 8; http://dx.doi.org/10.1145/602421.602426
- Kawano T, Bouteau F, Mancuso S. Finding and defining the natural automata acting in living plants: Towards the synthetic biology for robotics and informatics in vivo.. Commun Integr Biol 2012; In press http://dx.doi.org/10.4161/cib.21805
- Kawano T. Biomolecule-assisted natural computing approaches for simple polynomial algebra over fields. ICIC Exp Lett 2013; In press
- Yokawa K, Kagenishi T, Kawano T. Prevention of oxidative DNA degradation by copper-binding peptides. Biosci Biotechnol Biochem 2011; 75:1377 - 9; http://dx.doi.org/10.1271/bbb.100900; PMID: 21737913
- Yokawa K, Kadono T, Suzuki Y, Suzuki T, Uezu K, Kawano T. DNA-mediated sensitive detection and quantification of rare earth ions using polymerase chain reaction. Sens Mater 2011; 23:219 - 28