Abstract
Recently, we have reported the initial characterization of a novel centrin from Dictyostelium discoideum (DdCenB). Sequence and phylogenetic analyses clearly establish DdCenB as a centrin, yet further characterization revealed some interesting peculiarities about this novel centrin. Figure 1 depicts the localization of DdCenB at three points in the cell cycle: interphase, mitosis, and cytokinesis. In interphase DdCenB primarily localizes to the nuclear envelope (NE). Although the NE remains intact during mitosis and cytokinesis in Dictyostelium, DdCenB disappears from the NE at these two stages of the cell cycle. In addition to localization at the NE, we also see weak localization in the nucleoplasm and cytoplasm (weakest). Although the nucleoplasmic concentration appears constant throughout the cell cycle, the very faint localization in the cytoplasm does appear to increase to the level of the nucleoplasm during mitosis and cytokinesis. Unlike most centrins characterized to date, we found no evidence of DdCenB at the centrosome at any point in the cell cycle. Here we examine the importance of DdCenB localization in cell cycle progression, as well as several other roles.
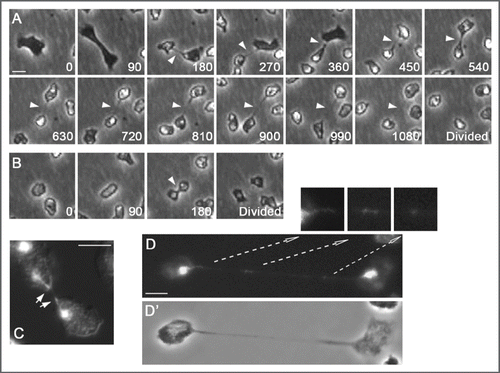
To further explore DdCenB we generated a knock-out strain. One of the first phenotypes we observed was a difficulty in completing cytokinesis. Unable to synchronize cells, we relied on time-lapse photomicroscopy to observe cells for prolonged periods of time. We commonly observed dividing cells that remained interconnected by structures resembling cytoplasmic bridges. These bridges often persisted for over 15 minutes (), even though Dictyostelium cells can typically use traction to complete cytokinesis.Citation2 This strongly contrasted with wild-type cells, which divide relatively rapidly () and without the formation of a true midbody.Citation3 Mitotic defects can often times lead to aberrations in cytokinesis. Consequently, we also examined DdCenB knock-out cells in mitosis. Indicative of mitotic defects, we observed aberrant mitotic spindles and mislocalization of the microtubule binding protein DdCP224, a centrosome protein that also localizes to the mitotic spindle.Citation4 Interestingly, altered expression of DdCP224 also generates mitotic and centrosome defectsCitation5 similar to those observed with DdCenB knock-out. In some cases, cells delayed in cytokinesis retained microtubule bundles in the connecting cytoplasmic bridges (presumably remaining from the mitotic spindle) and chromosomes delayed in reaching the poles (). These persistent microtubules and delayed chromosomes are hallmarks of spindle defects and it may be because of these defects that cytokinesis is impaired.
How can it be that defects in mitosis and cytokinesis result from the disruption (either through knock-out or overexpression) of a protein that apparently disappears at these stages of the cell cycle? Part of the answer may lie in what happens to the centrosome when DdCenB function is compromised. Immuno-staining of DdCenB knock-out cells with NE and centrosome markers revealed changes in the normal centrosome-nucleus association. In wild-type Dictyostelium cells the centrosome is closely tethered to the NE during interphase.Citation6 The lack of DdCenB led to a protrusion of the NE towards a displaced centrosome. This phenotype is similar to that obtained upon overexpression of full length or truncated DdSun1, a SUN domain protein with an established role in anchoring the centrosome to the nucleus.Citation7,Citation8 SUN domain proteins span the NE and ultimately link centrosomeinteracting KASH domain proteins to lamins (metazoans) or DNA (yeast, Dictyostelium).Citation9,Citation10 The similarity of phenotype between DdCenB knock-out and DdSun1 overexpressing cells suggests that DdCenB might also function in tethering the centrosome to the NE. In addition to defects in the centrosome-NE link, or perhaps because of this, we have also observed multiple centrosomes and centrosomerelated bodies in DdCenB knock-out cells. The presence of altered centrosomes would certainly have profound effects on the ability of those cells to establish a normal mitotic spindle and proceed through cell division in a normal fashion. Alternatively, DdCenB (before delocalizing from the NE) may aid in the transition of the centrosome into the NE in a similar fashion to yeast Cdc31 and the nascent spindle pole body (SPB).Citation11 Disruption of this process would release the centrosomes from the NE and allow for aberrant rounds of centrosome duplication.
Regarding cytokinesis, in addition to the possibility that these defects are simply a result of defects in mitosis, it is also possible that DdCenB actively participates in the regulation of cytokinesis. The mechanism by which DdCenB is lost from the NE as cells transition into mitosis has not been determined. It is possible that DdCenB, released from the NE at this time, becomes cytoplasmic and more directly involved in cytokinesis. Centrins are known to be calcium sensors and there is evidence for calcium in cytokinesis.Citation12 Since we have not observed a distinct subcellular localization for DdCenB in dividing cells (e.g., the cleavage furrow) it remains feasible that DdCenB acts more globally at this stage in the cell cycle. For example racE, a small GTPbinding protein required for cytokinesis in Dictyostelium, is necessary for maintaining proper cortical tension, essential for furrow completion.Citation13 Considering its involvement with the NE, DdCenB may play a role in membrane trafficking during cytokinesis.
The multiplicity of phenotypes observed following DdCenB knock out or overexpression, coupled with its peculiar cell cycle dependent localization suggests multiple roles for DdCenB. In addition to localization at the NE, there is faint localization in the nucleoplasm (see ). Unlike DdCenB at the nuclear envelope, the nucleoplasmic DdCenB remains there throughout the cell cycle. This suggests that nucleoplasmic DdCenB might constitute a different protein population with different roles. Reports have shown that centrins are required to stimulate DNA repair mechanisms. For example, yeast CDC31 and human centrin 2 bind and stimulate the NEF2 complex and XPC, respectively.Citation14 A homolog to human XPC has been identified in Dictyostelium. Consequently, nucleoplasmic DdCenB may play a similar role to yeast and human centrins in the stimulation of nucleotide excision repair mechanisms.
Figures and Tables
Figure 1 Localization of DdCenB at three stages of the cell cycle. DdCenB is shown as gray shading, the darkness of which corresponds to the amount of DdCenB. N, nucleus; NE, nuclear envelope; C, centrosome; PM, plasma membrane.
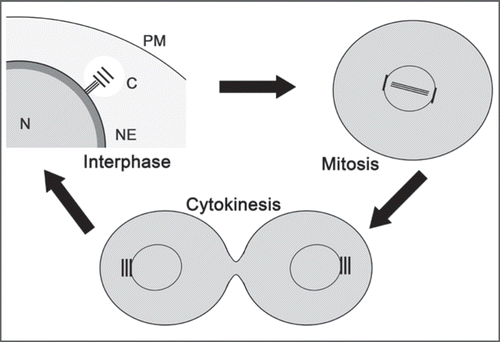
Figure 2 Cytokinesis defects as a result of DdCenB knock-out. (A) consists of images captured from time-lapse microscopy. The time of capture (minutes) for each frame is indicated in the lower right corners. The arrowheads point to the persistent cytoplasmic bridges observed in dividing cells. In (B) we see a wild-type cell dividing. Here the arrowhead points to a typical cleavage furrow. (C) is a dividing mutant cell stained with DA PI (DNA) and visualized by fluorescence/DIC microscopy. Arrows point to distorted nuclei in the furrow region. (D) shows a mutant cell, with a long cytoplasmic bridge, stained with DAPI. The arrows point to higher magnifications of the respective areas, and highlight the presence of DNA in the cytoplasmic bridge. D, fluorescence; D’, phase contrast. Bars in all cases correspond to 5 µm.
Addendum to:
References
- Mana-Capelli S, Gräf R, Larochelle DA. Dictyostelium discoideum CenB is a bona fide centrin essential for nuclear architecture and centrosome stability. Eukaryotic Cell 2009; 8:1106 - 1117
- Uyeda TQ, Nagasaki A, Yumura S. Multiple parallelisms in animal cytokinesis. Int Rev Cytol 2004; 240:377 - 432
- Chen Q, Li H, De Lozanne A. Contractile Ring-independent Localization of DdINCENP, a Protein Important for Spindle Stability and Cytokinesis. Mol Biol Cell 2006; 17:779 - 788
- Graf R, Daunderer C, Schliwa M. Dictyostelium DdCP224 is a microtubule-associated protein and a permanent centrosomal resident involved in centrosome duplication. J Cell Sci 2000; 113:1747 - 1758
- Graf R, Euteneuer U, Ho TH, Rehberg M. Regulated expression of the centrosomal protein DdCP224 affects microtubule dynamics and reveals mechanisms for the control of supernumerary centrosome number. Mol Biol Cell 2003; 14:4067 - 4074
- Graf R, Brusis N, Daunderer C, Euteneuer U, Hestermann A, Schliwa M, et al. Comparative structural, molecular and functional aspects of the Dictyostelium discoideum centrosome. Curr Topics Dev Biol 2000; 49:161 - 185
- Schulz I, Baumann O, Samereier M, Zoglmeier C, Graf R. Dictyostelium Sun1 is a dynamic membrane protein of both nuclear membranes and required for centrosomal association with clustered centromeres. Eur J Cell Biol 2009;
- Xiong H, Rivero F, Euteneuer U, Mondal S, Mana-Capelli S, Larochelle D, et al. Dictyostelium Sun-1 connects the centrosome to chromatin and ensures genome stability. Traffic 2008; 9:708 - 724
- Starr DA, Fischer JA. KASH ‘n Karry: the KASH domain family of cargo-specific cytoskeletal adaptor proteins. Bioessays 2 2005; 27:1136 - 1146
- Tzur YB, Wilson KL, Gruenbaum Y. SUN-domain proteins: ‘Velcro’ that links the nucleoskeleton to the cytoskeleton. Nat Rev Mol Cell Biol 2006; 7:782 - 788
- Spang A, Courtney I, Fackler U, Matzner M, Schiebel E. The calcium-binding protein cell division cycle 31 of Saccharomyces cerevisiae is a component of the half bridge of the spindle pole body. J Cell Biol 1993; 123:405 - 416
- Moncrief ND, Kretsinger RH, Goodman M. Evolution of EF-hand calcium-modulated proteins. I. Relationships based on amino acid sequences. J Mol Evol 1990; 30:522 - 562
- Larochelle DA, Gerald N, De Lozanne A. Molecular analysis of racE function in Dictyostelium. Microsc Res Tech 2000; 49:145 - 151
- Chen L, Madura K. Centrin/Cdc31 is a novel regulator of protein degradation. Mol Cell Biol 2008; 28:1829 - 1840