Abstract
Both the Rho GTPases and 14-3-3 proteins each belong to ubiquitous families of proteins involved in a variety of cellular processes, including cytokinesis, cell polarity, cellular differentiation, and apoptosis. In fungi, these components of signaling pathways are involved in cell cycle regulation, cytokinesis, and virulence. We study cellular differentiation and pathogenesis for Ustilago maydis, the dimorphic fungal pathogen of maize. We have reported on the interactions of Pdc1, a U. maydis homologue of human 14-3-3ε, with Rho1, a small GTP binding protein; these proteins participate in cell polarity and filamentation pathways that include another small G protein, Rac1, and its effector PAK kinase, Cla4. Here we describe additional experiments that explore possible relationships of Pdc1 and Rho1 with another PAK-like kinase pathway and with the a mating-type locus.
Two highly-conserved signaling pathways are present in a variety of fungi and many of their components are conserved from fungi to humans.Citation1 These pathways, the MAPK and cAMP-dependent PKA pathways, play roles in cellular differentiation, responses to environmental signals such as nutrient levels and stress, response to pheromone for mating, and virulence.Citation2
The 14-3-3 proteins and the Rho small GTP-binding proteins are ubiquitous in eukaryotic cells as regulators of these and other signaling pathways and each belongs to a large family of related proteins. The 14-3-3 proteins are members of a large family of highly conserved and ubiquitously-expressed proteins. Seven isotypes are present in mammals and up to fifteen different isotypes have been discovered in plants.Citation3 Like Drosophila melanogaster and Caenorhabditis elegans,Citation4 fungi tend to have two isotypes, though both Candida albicansCitation5 and U. maydisCitation6 each have only a single gene encoding their respective 14-3-3 homologues. Members of this protein family of small, ∼30 kDa proteins, typically form dimers with one another to generate an active ligand-binding protein.Citation7
Their exact roles in the various cellular processes with which 14-3-3 proteins have been associated has been difficult to ascertain due to multiple isotypes per cell and the fact that over 200 interacting partners have been identified for 14-3-3 proteins.Citation8 Thus, in U. maydis we were provided with a unique opportunity to investigate the role(s) of 14-3-3 proteins, since only one homologue was predicted from inspection of the genome sequence. We identified a 14-3-3ɛ homologue in U. maydis, which we called Pdc1 (phosphorylationdomain coupling protein) since 14-3-3 dimers couple phosphoserine/threonine target proteins.Citation9 U. maydis Pdc1 is predicted to have 77% and 75% identity to Saccharomyces cerevisiae Bmh1 and Bmh2, respectively. In fact, when overexpressed in a S. cerevisiae Δbmh1bmh2 mutant, Pdc1 partially rescues the growth defect of the yeast mutant, suggesting at least some functional similarity as well.Citation6
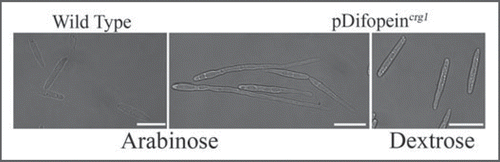
It was subsequently determined that Pdc1Citation10 plays a role in cell cycle control. However, in contrast to the role of its S. cerevisiae homologue, Bmh1, U. maydis Pdc1/Bmh1 was found to be involved in G2 transition, a function dependent on the phosphorylation state of the dualspecificity phosphatase, Cdc25.Citation10 Results from overexpression of a synthetic peptide inhibitor of 14-3-3 proteins, difopein,Citation11 seems to lend further support for the role of Pdc1 in cell cycle control. Cells expressing difopein, a competitive inhibitor that binds to the ligand-binding groove of 14-3-3 proteins, undergo polar growth extension () in a fashion similar to b-mating locus-induced filament formation. However, unlike b-mating locus-induced filament, the apical-constricted growth extensions induced by treatment of cells with difopein appear tapered at the active growing end. Such extensions are consistent with failure of cells to undergo cell division. In addition to its role in cell cycle control, Pdc1 also is required for viability in haploid cells and plays critical roles in cytokinesis, cell polarity and filamentation.Citation6
Rho (Ras homologue) GTPases are small molecular weight G-proteins belonging to the Ras superfamily, whose activity is dependent on the reversible binding of guanosine triphosphate and guanosine diphosphate.Citation12 They are involved in actin organization, polarized cell growth, cytokinesis and regulation of cell cycle.Citation13 In these processes, they play instrumental role(s) in establishing and maintaining cell polarity. Members of the Rho-family GTPases include Rho, Rac and Cdc42. In U. maydis, Rho1 is the only member of this family identified to date that is required for cell viability in haploid cells,Citation6 as its deletion or depletion leads to cell death. The phenotypes following its depletion or overexpression mirror those of Pdc1, with the added observation of reduced mating of U. maydis cells when Rho1 is overexpressed in the mating partners.Citation6 In addition, epistasis analyses suggest that both Pdc1 and Rho1 are upstream of cytokinesis and cell polarity pathways, ultimately regulated by another small G protein, Rac1, and its effector kinase, Cla4 [a p21-activated kinase (PAK)].Citation14,Citation15
In additional experiments with both Pdc1 and Rho1, we sought to explore further the interactions between these proteins and other signaling components affecting cell morphology and differentiation. U. maydis, like other fungi, has several homologues of PAK-like kinases.Citation16,Citation17 In particular, it contains a Cla4 homologue and a homologue of the yeast Ste20p, Smu1.Citation18 While neither of these kinases is required for viability, presumably due to some redundancy, at least in yeasts, deletion of both is lethal, suggesting overlap in function.Citation19
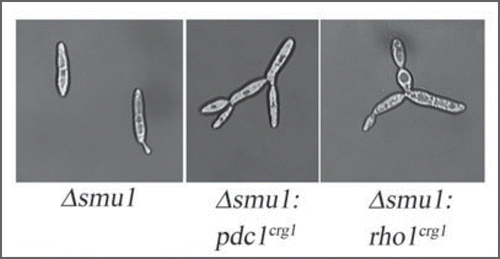
In S. cerevisiae, the 14-3-3 homologues, Bmh1 and Bmh2 regulate pseudohyphal development by binding to the p21-activated kinase Ste20 of the MAPK signaling pathway.Citation20 Thus, we were initially curious to examine similar potential roles for Pdc1 and/or Rho1 with regard Smu1 in U. maydis. As seen in , when either Pdc1 or Rho1 was overexpressed in a haploid strain deleted for the smu1 gene, we observed a multiple-budding phenotype, reminiscent of the ubc1 mutants that lack the regulatory subunit of PKA and thus have overactive catalytic subunit of the kinase.Citation21 No such phenotype was observed in wild type cells when these genes were overexpressed, although overexpression of pdc1 in wild type led to slight elongation of the cells.
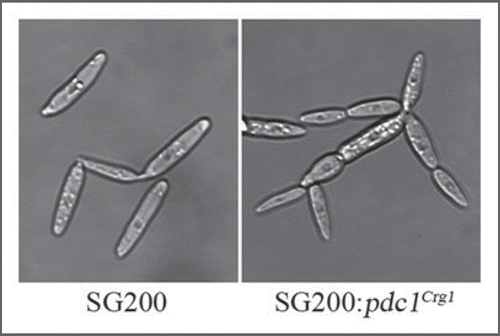
Our previous work with Pdc1 and Rho1 indicated that their role is likely upstream of the a and b mating-type loci.Citation6 In U. maydis the a locus encodes pheromone and receptors that control the cell-cell recognition that serves as a prelude to cell fusion; normally such fusion is a prerequisite for subsequent filamentation and infectivity, in turn controlled by the b locus.Citation22,Citation23 In genetically engineered strains where the cell fusion step has been bypassed, once filamentation has been initiated, neither depletion nor overexpression of Pdc1 or Rho1 has any observable effects on filamentation, although their depletion is eventually lethal.Citation6 The otherwise haploid SG200 strainCitation24 has been engineered so that it is heterozygous at the b locus and so that it produces autocrine signaling of pheromone to the corresponding receptor. This leads to upregulation of pheromone gene expression and then to filamentous growth under control of the b locus. When Pdc1 were overexpressed in SG200, the result was both multiplebudding and clusters of cells resembling “balloon dachshunds” ().
Here, we have added to our previously published analyses by providing (1) additional evidence of the role of Pdc1 in cell cycle (); (2) evidence that Pdc1 and Rho1 participate in the same or a parallel pathway with Smu1 (); and (3) data showing a possible role of Pdc1 in phenotypes associated with expression of the a locus (). We showed previouslyCitation18 that Smu1 is required for upregulation of the mfa gene in a2 cells upon exposure to pheromone from a1 cells. When a2 cells are deleted for smu1, they show reduced mating in both charcoal and confrontation mating assays, and they fail to increase mfa2 expression in such assays. Interestingly, based on yeast two-hybrid and co-immunoprecipitation analyses, Smu1 is predicted to interact with Rho1.Citation6 If so, this would suggest a role for Rho1 (and its upstream partner, Pdc1) in cell morphology associated with expression levels of the a locus.
Figures and Tables
Figures 1 Inhibition of Pdc1 activity by the synthetic peptide inhibitor, difopein, leads to elongated cells. The Pcrg1-controlled difopein expression-construct is integrated into the genome at the ip locus.Citation25 Arabinose was used for the induction of difopein production and this leads to cell elongation as Pdc1 is inhibited. In contrast, in dextrose, when difopein is not expressed, cells appear like wild type cells growing in arabinose-containing media. Scale bar = 10 µm.
Figures 2 Overexpression of rho1 or pdc1 in the absence of Smu1 leads to multiple-bud morphology. Deletion of smu1 does not cause any discernable phenotype (Δsmu1). Interestingly, when we overexpressed either pdc1 or rho1 in the smu1 background, the cells appeared as aggregates. The promoters of pdc1 and rho1 were replaced with the Pcrg1,Citation6 so that they would be overexpressed in the presence of arabinose.
Figures 3 Overexpression of pdc1 in SG200 cells leads to cell separation defect. SG200 cells are typically cigar-shaped. However, when we overexpressed pdc1 in this cell line, the cells appear highly branched. The promoter of pdc1 was replaced with the Pcrg1.Citation6 In arabinose-containing media the gene would be overexpressed.
Addendum to:
References
- Bardwell L. A walk-through of the yeast mating pheromone response pathway. Peptides 2005; 26:339 - 350
- Kelly MN, Johnston DA, Peel BA, Morgan TW, Palmer GE, Sturtevant JE. Bmh1p (14-3-3) mediates pathways associated with virulence in Candida albicans. Microbiology 2009; 155:1536 - 1546
- Mhawech P. 14-3-3 proteins-an update. Cell Research 2005; 15:228 - 236
- van Hemert J, Steensma HY, van Heusden GPH. 14-3-3 proteins: key regulators of cell division, signalling and apoptosis. BioEssays 2001; 23:936 - 946
- Palmer GE, Johnson KJ, Ghosh S, Sturtevant J. Mutant alleles of the essential 14-3-3 gene in Candida albicans distinguish between growth and filamentation. Mol Microbiol 2004; 150:1911 - 1924
- Pham CD, Yu Z, Sandrock B, Bölker M, Gold SE, Perlin MH. Ustilago maydis Rho1 and 14-3-3 homologues participate in pathways controlling cell separation and cell polarity. Eukaryot Cell 2009; 8:977 - 989
- Darling DL, Yingling J, Wynshaw-Boris A. Role of 14-3-3 proteins in eukaryotic signaling and development. Curr Top Dev Biol 2005; 68:281 - 315
- Kakiuchi K, Yamauchi Y, Taoka M, Iwago M, Fujita T, Ito T, et al. Proteomic analysis of in vivo 14-3-3 interactions in the yeast Saccharomyces cerevisiae. Biochemistry 2007; 46:7781 - 7792
- Fu H, Subramanian RR, Masters SC. 14-3-3 proteins: structure, function and regulation. Annu Rev Pharmacol Toxicol 2000; 40:617 - 647
- Mielnichuk N, Pérez-Martín J. 14-3-3 regulates the G2/M transition in the basidiomycete Ustilago maydis. Fungal Genet Biol 2008; 45:1206 - 1215
- Masters SC, Fu H. 14-3-3 Proteins mediate an essential anti-apoptotic signal. J Biol Chem 2001; 276:45193 - 451200
- Narumiya S. The small GTPase Rho: cellular functions and signal transduction. J Biochem 1996; 120:215 - 228
- Jaffe A, Hall A. Rho GTPases: biochemistry and biology. Annu Rev Cell Dev Biol 2005; 21:247 - 269
- Leveleki L, Mahlert M, Sandrock B, Bölker M. The PAK family kinase Cla4 is required for budding and morphogenesis in Ustilago maydis. Mol Microbiol 2004; 54:396 - 406
- Mahlert M, Leveleki K, Hlubek A, Sandrock B, Bölker M. Rac1 and Cdc42 regulate hyphal growth and cytokinesis in the dimorphic fungus Ustilago maydis. Mol Microbiol 2006; 59:567 - 578
- Klosterman SJ, Perlin MH, Garcia-Pedrajas M, Covert SF, Gold SE. Genetics of morphogenesis and pathogenic development of Ustilago maydis. Adv Genet 2007; 57:1 - 47
- García-Pedrajas MD, Nadal M, Bölker M, Gold SE, Perlin MH. Sending mixed signals: Redundancy vs. uniqueness of signaling components in the plant pathogen, Ustilago maydis. Fungal Genet Biol 2008; 45:22 - 30
- Smith DG, Garcia-Pedrajas MD, Hong W, Yu Z, Gold SE, Perlin MH. An ste20 homologue in Ustilago maydis plays a role in mating and pathogenicity. Eukaryot Cell 2004; 3:180 - 189
- Keniry ME, Sprague GF Jr. Identification of p21-activated kinase specificity determinants in budding yeast: a single amino acid substitution imparts Ste20 specificity to Cla4. Mol Cell Bio 2003; 23:1569 - 1580
- Roberts RL, Mösch HU, Fink GR. 14-3-3 Proteins are essential for RAS/MAPK cascade signaling during pseudohyphal development in S. cerevisiae. Cell 1997; 89:1055 - 1065
- Gold S, Duncan G, Barrett K, Kronstad J. cAMP regulates morphogenesis in the fungal pathogen Ustilago maydis. Genes Dev 1994; 8:2805 - 2816
- Banuett F, Herskowitz I. Different a alleles of Ustilago maydis are necessary for maintenance of filamentous growth but not for meiosis. Proc Natl Acad Sci USA 1989; 86:5878 - 5882
- Bölker M. Ustilago maydis—a valuable model system for the study of fungal dimorphism and virulence. Microbiology 2001; 147:1395 - 1401
- Bölker M, Genin S, Lehmler C, Kahmann R. Genetic regulation of mating and dimorphism in Ustilago maydis. Can J Bot 1995; 73:320 - 325
- Brachmann A, Weinzierl G, Kämper J, Kahmann R. Identification of genes in the bW/bE regulatory cascade in Ustilago maydis. Mol Microbiol 2001; 42:1047 - 1063