Abstract
A key feature of Hodgkin lymphoma is that the malignant cells are binucleated, as a consequence of failed cytokinesis. We recently ascertained a family in which multiple cases of Hodgkin lymphoma had occurred among individuals who inherited a balanced chromosomal translocation. We cloned the translocation breakpoints and found that it disrupted a previously uncharacterized gene, KLHDC8B, encoding a Kelch family protein whose deficiency impairs cytokinesis and leads to binucleated cells. In other families we found a rare single nucleotide polymorphism affecting mitotic translation of KLHDC8B that was associated with and linked to Hodgkin lymphoma. Interestingly, the index family demonstrated an unusual frequency of twins, and there is a previously reported association between Hodgkin lymphoma and twins. Here we review the unusual genetic features of Hodgkin lymphoma, including gender concordance among siblings, and genetically test the hypothesis that KLHDC8B may participate in twinning by disrupting cytokinesis through impediment of polar body separation from oocytes.
Hodgkin lymphoma is a lymph node cancer of B-cell origin.Citation1 An important feature is that in classical Hodgkin lymphoma, the malignant cells, known as “Reed-Sternberg” cells, are typically binucleated. In contrast to other cancers, where the majority of the cells comprising the tumor appear to be malignant, Reed-Sternberg cells are few in number and are surrounded by a sea of reactive, yet benign, inflammatory cells.
Of all types of malignancy, Hodgkin lymphoma demonstrates particularly strong heritability, in that relatives of cases are at an especially high risk for also developing the disease, compared to many other types of cancer.Citation2 However, except for HLA associationsCitation3 and rare autoimmune lymphoproliferative disordersCitation4 whose clinical features include other problems, no genes predisposing to development of Hodgkin lymphoma in the general population had been recognized.
Gender Concordance in Hodgkin Lymphoma
We had previously proposed a pseudoautosomal locus for Hodgkin lymphoma.Citation5,Citation6 A curious phenomenon is that when Hodgkin lymphoma runs within families, affected individuals tend to be of the same sex.Citation2,Citation7 Among sibling pairs with Hodgkin lymphoma, male-male or female-female pairs predominate; male-female pairs are fewer. The most striking example (no pun intended), occurs in what is perhaps the largest Hodgkin disease kindred ever described, that of New York Yankee baseball legend, Mickey MantleCitation8 (). In his family (), six people developed Hodgkin lymphoma, and a seventh developed non-Hodgkin lymphoma. (Mickey himself succumbed to hepatocellular carcinoma). All were male. Pseudoautosomal genetic transmission could explain gender concordance in a pedigree such as the Mantle family, because the pseudoautosomal regions contain identical genes on both sex chromosomes. Should a pseudoautosomal mutation derive from the father, then it could reside on either the X or Y chromosome and, recombination notwithstanding, would necessarily follow gender. A supportive observation is that Hodgkin lymphoma has been observed to co-segregate in a pedigreeCitation9,Citation10 transmitting the only known genetic disorder (Leri-Weil dyschondrosteosis) attributed to a gene (SHOX) residing within the pseudoautosomal regions of the sex chromosomes. To date, though, this hypothesis has not been rigorously evaluated and remains conjectural.
KLHDC8B in Hodgkin Lymphoma
Nevertheless, our laboratory has maintained interest in Hodgkin lymphoma and has continued to seek rare families with multiple occurrences in the hope that studying them might illuminate the intriguing genetic principles underlying this form of cancer. Recently, a family presented to us with multiple cases of the nodular sclerosis type of classical Hodgkin lymphoma and other malignancies.Citation11 Interestingly, all individuals with cancer for whom genetic material was available carried a constitutional, balanced translocation involving chromosomes 2 and 3.
In the hopes that the translocation could point to a potential tumor suppressor gene, we molecularly cloned the translocation and determined that it indeed disrupted a previously uncharacterized gene, KLHDC8B, located at a site of recurrent cytogenetic abnormalities and loss of heterozygosity (LOH) in B-cell lymphomas12 and other malignanciesCitation13,Citation14 on chromosome 3. This region has also been implicated in nasopharyngeal carcinoma,Citation15–Citation17 which, like Hodgkin lymphoma,Citation1 shares an association with Epstein-Barr virus.Citation18 We additionally found that a rare 5′-UTR SNP, which appears to alter mitotic-specific expression of KLHDC8B, was both associated with and linked to Hodgkin lymphoma in three more families. Further, in one of three sporadically occurring cases of Hodgkin lymphoma, we detected somatic LOH for KLHDC8B in Reed-Sternberg cells, but not in reactive T-lymphocytes purified from the tumor.
We developed an antibody to KLHDC8B and determined that the protein is expressed only during cytokinesis and locates to the midbody, which is a structure that connects dividing cells just prior to their separation. RNAi knockdown of KLHDC8B resulted in an increase in binucleated cells. When cytokinesis cannot be completed, the cleavage furrow regresses, culminating in a binucleated cell. Thus, deficiency of KLHDC8B recapitulates the signature Reed-Sternberg cell of Hodgkin lymphoma.
KLHDC8B in Twinning
KLHDC8B encodes a protein predicted to contain seven repeated “Kelch” domains.Citation19 The Kelch domain was discovered in a Drosophila protein component of ring canals, which form syncytia interconnecting oocytes and which derive from incomplete cytokinesis of primordial germ cells.Citation20 Other mammalian homologs of Kelch, including KLHL9 and KLHL13, also locate to the midbody and produce binucleated cells when knocked-down by RNAi.Citation21 In mammals, oocytes similarly develop from “oocyte nests”—clusters of primordial germ cells joined through ring canals that break down to release individual eggs.Citation22 Gametogenesis is therefore coordinated with cytokinesis.
Interestingly, the index familyCitation11 segregating Hodgkin lymphoma with the chromosomal translocation disrupting KLHDC8B had three sets of apparent dizygotic twins occurring among eight births. We therefore entertained the hypothesis that KLHDC8B may contribute to twinning, as well as Hodgkin lymphoma. It is conceivable that disruption of KLHDC8B may lead to production of pairs of gametes tethered to one another through a persistent cytoplasmic bridge, akin to the physiologic role of Kelch in Drosophila. Along these lines, it is worth noting that the chromosomal region containing KLHDC8B was among top-scoring loci in a genome-wide linkage scan for dizygotic twinning.Citation23 Moreover, rather uniquely among different forms of cancer, being born as a dizygotic twin is associated with an increased risk for Hodgkin lymphoma,Citation24 suggesting that there may be a common genetic basis for both.
We specifically weighed the possibility that a deficiency of cytokinesis may impair dissociation of an oocyte from a polar body, during either first or second meiosis, thus resulting in the phenomenon of “polar body twins”. (Polar bodies are byproducts of female gametogenesis not maturing to oocytes but that are nonetheless genetically equivalent to oocytes). While there is only scant evidence that polar body twinning has ever actually occurred,Citation25 twins derived from an ovum and its first polar body would be expected to be less genetically similar to one another than ordinary siblings or dizygotic twins, while twins derived from an ovum and its second polar body are postulated to be intermediate in genetic identity between dizygotic twins (or ordinary siblings) and monozygotic twins.Citation26,Citation27
The genetic relationship between polar body twins, if they do really occur, would be determined by two major factors: which polar body undergoes fertilization and the number and distribution of chiasmata during oogenesis. The first polar body is created after the initial stage of meiosis and therefore is necessarily genetically distinct from the secondary oocyte; however, the second polar body is derived from the secondary oocyte so is genetically identical ().
In the absence of chiasmata it should be relatively straightforward to discriminate between first and second polar body twins. If a derivative of the first polar body were subsequently fertilized, then all maternally derived genes would be necessarily non-identical. However, if the second polar body is fertilized then all maternally derived genes will be identical.
This is complicated significantly by crossing over during meiosis. At an individual locus, if no chiasmata have occurred, then the assumption outlined in the previous paragraph is correct. A single chiasma, however, would result in all-maternally derived alleles telemeric of the breakpoint being non-identical in second polar body twins. In contrast, for first polar body twins, a single chiasma would result in a 50% chance of maternally derived alleles telemeric of the breakpoint being identical (and a 50% chance of being non-identical) (). This is complicated further when two or more chiasmata occur.
The overall expectation for sharing in first and second polar body twins was examined by Goldgar and Kimberling.Citation26 In summary, the correlation between first and second polar body twins is expected to be 38 and 51%, respectively. Using a set of markers spread throughout the genome and calculating overall allele sharing would permit identification of first polar body twins, but would not be sufficient to distinguish second polar twins from dizygotic twins. These limitations could be overcome in two ways: either by mapping all chiasmata or by using markers adjacent to the centromere to reconstruct allele sharing in the absence of chiasmata.
We selected one pair of twins from the pedigree, whose mother had the translocation disrupting KLHDC8B and who developed Hodgkin lymphoma. Each of the twins was male, neither inherited the translocation nor has had Hodgkin lymphoma, and they were considered by their family to be non-identical. To examine their relationship we undertook a whole genome screen using the Genechip Mapping 10K Array (Affymetrix), containing approximately 10,200 SNPs.
We initially determined overall correlation using the program PLINK.Citation28 This demonstrates an overall sharing of 54%, with sharing of 0, 1 and 2 alleles being 21, 49 and 30%, respectively (compared to an expected distribution of 25, 50 and 25%). This indicates that it is unlikely that the twins were first polar body twins. However, by analyzing overall sharing statistics we can not rule out the twins being second polar body twins.
To explore the relationship between the twins further, we analyzed identity by descent (IBD) sharing using the programs MERLINCitation29 and RELATE.Citation30 Probabilities of sharing 0, 1 and 2 alleles were graphed, and the most probable pattern of sharing at centromeric regions was established (). The two programs gave similar results, with both indicating that the twins examined in this study are most likely dizygotic.
Overall, this analysis does not suggest that there is anything genetically special about this particular pair of twins. Nevertheless, the results are not definitive, because, in the absence of parental genotypes, allele sharing will be overestimated due to fortuitous sharing of common markers between both parents. Nor can we exclude that a disruption of cytokinesis might contribute to twinning through a mechanism other than involving ejection of polar bodies. For example, one could imagine that primordial germ cells remain connected, such that two primary oocytes may be deposited into a follicle and then ovulated simultaneously. The possibility that twinning and gender concordance could somehow be coupled (noting that there is a pair of twins in the Mantle family) through a mechanism other than that involving pseudoautosomal inheritance should also not be overlooked. We therefore plan to further explore the link between KLHDC8B, failed cytokinesis, and the curious genetic phenomena of twinning and gender concordance that have been associated with Hodgkin lymphoma.
Figures and Tables
Figure 1 Gender concordance in familial Hodgkin lymphoma. (A) Mickey Mantle and his four sons. (Photo courtesy of Danny Mantle). (B) Pedigree of the Mantle family. Hodgkin (HL, black) or non-Hodgkin lymphoma (NHL, grey). HCC, hepatocellular carcinoma.
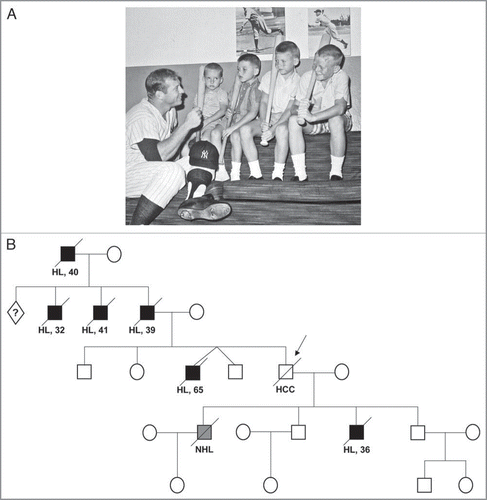
Figure 2 Graphical representation of polar body twinning and its implication in allele sharing. We compare allele sharing in first and second polar body twins (twin B) with a conventionally conceived twin (twin A). In each twin there is a 50% chance of sharing paternally derived alleles (represented by light and dark blue). However, the sharing of maternally derived alleles (represented by red and pink) is determined by which polar body is fertilized and the location and frequency of chiasmata. (Crossing-over is not depicted during spermatogenesis).
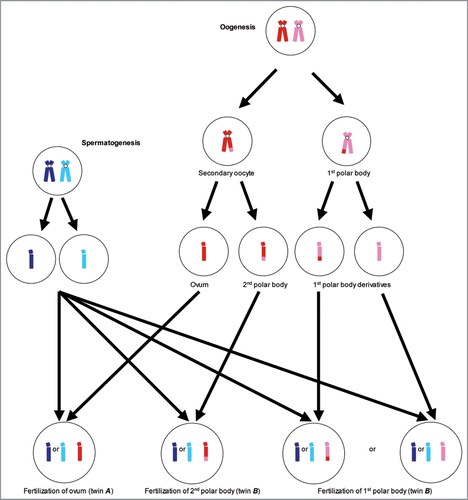
Table 1 Expected and observed allele sharing at centromeric regions throughout the genome indicative of twin type
Acknowledgements
AET is supported by NIH T32GM007454. We thank the families for their participation.
Addendum to:
References
- Kuppers R. The biology of Hodgkin’s lymphoma. Nat Rev Cancer 2009; 9:15 - 27
- Altieri A, Hemminki K. The familial risk of Hodgkin’s lymphoma ranks among the highest in the Swedish Family-Cancer Database. Leukemia 2006; 20:2062 - 2063
- Diepstra A, Niens M, te Meerman GJ, Poppema S, van den Berg A. Genetic susceptibility to Hodgkin’s lymphoma associated with the human leukocyte antigen region. Eur J Haematol Suppl 2005; 34 - 41
- Rieux-Laucat F, Le Deist F, Fischer A. Autoimmune lymphoproliferative syndromes: genetic defects of apoptosis pathways. Cell Death Differ 2003; 10:124 - 133
- Horwitz M, Wiernik PH. Pseudoautosomal linkage of Hodgkin disease. Am J Hum Genet 1999; 65:1413 - 1422
- Horwitz MS, Mealiffe ME. Further evidence for a pseudoautosomal gene for Hodgkin’s lymphoma: Reply to ‘The familial risk of Hodgkin’s lymphoma ranks among the highest in the Swedish Family- Cancer Database’ by Altieri A and Hemminki K. Leukemia 2 2007; 21:351
- Grufferman S, Cole P, Smith PG, Lukes RJ. Hodgkin’s disease in siblings. N Engl J Med 1977; 296:248 - 250
- Anderson D. Sports of the Times; on being Mickey Mantle. New York Times. New York 1994; 11
- Gokhale DA, Evans DG, Crowther D, Woll P, Watson CJ, Dearden SP, et al. Molecular genetic analysis of a family with a history of Hodgkin’s disease and dyschondrosteosis. Leukemia 1995; 9:826 - 833
- Shears DJ, Endris V, Gokhale DA, Dearden SP, Radford JA, Rappold GA, Taylor GM. Pseudoautosomal linkage of familial Hodgkin’s lymphoma: molecular analysis of a unique family with Leri-Weill dyschondrosteosis and Hodgkins lymphoma. Br J Haematol 2003; 121:377 - 379
- Salipante SJ, Mealiffe ME, Wechsler J, Krem MM, Liu Y, Namkoong S. Mutations in a gene encoding a midbody kelch protein in familial and sporadic classical Hodgkin lymphoma lead to binucleated cells. Proc Natl Acad Sci USA 2009; 106:14920 - 14925
- Szeles A. Fluorescence in situ hybridization (FISH) in the molecular cytogenetics of cancer. Acta Microbiol Immunol Hung 2002; 49:69 - 80
- Yang Q, Yoshimura G, Mori I, Sakurai T, Kakudo K. Chromosome 3p and breast cancer. J Hum Genet 2002; 47:453 - 459
- Hesson LB, Cooper WN, Latif F. Evaluation of the 3p21.3 tumour-suppressor gene cluster. Oncogene 2007; 26:7283 - 7301
- Xiong W, Zeng ZY, Xia JH, Xia K, Shen SR, Li XL, et al. A susceptibility locus at chromosome 3p21 linked to familial nasopharyngeal carcinoma. Cancer Res 2004; 64:1972 - 1974
- Cheng Y, Poulos NE, Lung ML, Hampton G, Ou B, Lerman MI, Stanbridge EJ. Functional evidence for a nasopharyngeal carcinoma tumor suppressor gene that maps at chromosome 3p21.3. Proc Natl Acad Sci USA 1998; 95:3042 - 3047
- Chan AS, To KF, Lo KW, Mak KF, Pak W, Chiu B, et al. High frequency of chromosome 3p deletion in histologically normal nasopharyngeal epithelia from Southern Chinese. Cancer Res 2000; 60:5365 - 5370
- Kutok JL, Wang F. Spectrum of Epstein-Barr virus-associated diseases. Annu Rev Pathol 2006; 1:375 - 404
- Prag S, Adams JC. Molecular phylogeny of the kelch-repeat superfamily reveals an expansion of BTB/kelch proteins in animals. BMC Bioinformatics 2003; 4:42
- Xue F, Cooley L. kelch encodes a component of intercellular bridges in Drosophila egg chambers. Cell 1993; 72:681 - 693
- Sumara I, Quadroni M, Frei C, Olma MH, Sumara G, Ricci R, Peter M. A Cul3-based E3 ligase removes Aurora B from mitotic chromosomes, regulating mitotic progression and completion of cytokinesis in human cells. Dev Cell 2007; 12:887 - 900
- Pepling ME, Spradling AC. Mouse ovarian germ cell cysts undergo programmed breakdown to form primordial follicles. Dev Biol 2001; 234:339 - 351
- Derom C, Jawaheer D, Chen WV, McBride KL, Xiao X, Amos C. Genome-wide linkage scan for spontaneous DZ twinning. Eur J Hum Genet 2006; 14:117 - 122
- Hemminki K, Li X. Cancer risks in twins: results from the Swedish family-cancer database. Int J Cancer 2002; 99:873 - 878
- Bieber FR, Nance WE, Morton CC, Brown JA, Redwine FO, Jordan RL, Mohanakumar T. Genetic studies of an acardiac monster: evidence of polar body twinning in man. Science 1981; 213:775 - 777
- Goldgar DE, Kimberling WJ. Genetic expectations of polar body twinning. Acta Genet Med Gemellol (Roma) 1981; 30:257 - 266
- Elston RC, Boklage CE. An examination of fundamental assumptions of the twin method. Prog Clin Biol Res 1978; 24:189 - 199
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007; 81:559 - 575
- Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin—rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet 2002; 30:97 - 101
- Albrechtsen A, Sand Korneliussen T, Moltke I, van Overseem Hansen T, Nielsen FC, Nielsen R. Relatedness mapping and tracts of relatedness for genome-wide data in the presence of linkage disequilibrium. Genet Epidemiol 2009; 33:266 - 274