Abstract
A key regulator of the slow recycling of receptors and lipids that occurs from the endocytic recycling compartment (ERC) back to the cell surface is EHD1. We have recently identified the Rab8a-interacting protein, MICAL-L1, as a novel binding partner for EHD1 that both recruits and interacts with EHD1 on tubular recycling endosomes. MICAL-L1 belongs to the MICAL-family of proteins that are highly expressed in neurons and involved in plexin-mediated repulsive axon guidance. Interestingly, MICAL-L1 contains a coiled coil region in its C-terminus that is both necessary and sufficient for its localization to the EHD1-containing long tubular membranes of the ERC. Furthermore, MICAL-L1-depletion also impaired recycling of both transferrin and integrin receptors from the ERC back to the plasma membrane. In conclusion, our studies implicate MICAL-L1 as a novel regulator of endocytic recycling, and raises the possibility that additional neuronal-expressed proteins may mediate endocytic events in non-neuronal cells.
The plasma membrane is a dynamic interface that constantly communicates with its surrounding external environment and whose composition is highly regulated in order to generate appropriate responses to extracellular cues. A dynamic interplay of internalization and recycling events controls the surface area and composition of the plasma membrane while maintaining a constant flux of material in and out of the cell. Although much is known about the cellular machinery mediating internalization of receptor-ligand complexes, somewhat less is known about the mechanisms by which some internalized cargo molecules are recycled back to the cell surface. Recycling can occur directly from the early endosomes by a ‘fast recycling’ pathway; however, most receptors are first transported to a compartment known as the endocytic recycling compartment (ERC) from where they traffic via a ‘slow-recycling’ route back to the cell surface.Citation1,Citation2 The ERC, in several cell types, is a series of tubular and vesicular membranes that appear to emanate from the microtubule organizing center and is defined by the presence of proteins such as Rab11,Citation3 and more recently EHD1, a member of C-terminal EHD family of proteins (reviewed in ref. Citation4).
EHD1 localizes to the cytosolic face of long tubular membranes of the ERC from where it mediates recycling of various transmembrane receptors including transferrin receptor, MHC class I, insulinresponsive glucose transporter (GLUT4) and β1 integrinsCitation5–Citation9 and others (reviewed in ref. Citation4). A recent study by Jovic et al. (2009) has shown that EHD1 localization to tubular membranes of the ERC is necessary for efficient receptor recycling to the plasma membrane.Citation10 However, a key unresolved question was whether EHD1 induces generation of these tubular structures, or decorates pre-existing membranes. In vitro studies using purified liposomes have clearly demonstrated the ability of EHD proteins to generate tubules.Citation11 In vivo, however, these tubules are much wider and proteins that colocalize with EHD1-containing tubules such as Rab8a,Citation12 continue to associate with tubular membranes even in the absence of EHD1, suggesting that EHD1 associates with pre-existing tubular membranes.Citation10
To elucidate the mechanism by which EHD1 is recruited to ERC tubular membranes, we performed GST-pulldown experiments using the EH-domain of EHD1. By mass spectrometry analysis, we identified a member of MICAL-family of proteins known as Molecule interacting with CasL (MICAL)-like 1 (MICAL-L1), as a novel interaction partner of EHD1.Citation13 MICAL proteins were originally identified as novel binding partners for CasL/HEF1, a protein that primarily localizes to focal adhesions and plays crucial role in integrin-induced signaling.Citation14 MICALs are large multidomain-containing proteins that are expressed primarily in neurons, but also in various non-neuronal cell types. In neurons, MICALs have been shown to bind plexins, receptors for semaphorins involved in mediating repulsive motor axon guidance.Citation15 An intact N-terminal flavoprotein monooxygenase domain (FAD) of MICAL was required for mediating plexin signaling. Interestingly, MICALlike proteins (MICAL-L1 and MICAL-L2) do not contain a FAD domain suggesting distinct functions for these proteins. In accordance with this, MICAL-L2 has been shown to regulate recycling of tight junction and adherens junction proteins back to the plasma membrane, while the role of MICAL-L1 has remained unaddressed so far. Interestingly, MICAL-L1 is the only MICAL family member that contains two NPF tripeptide motifs; sequences that bind to EH-domains, a domain characteristic of EHD1 and various proteins involved in endocytic trafficking (see ).
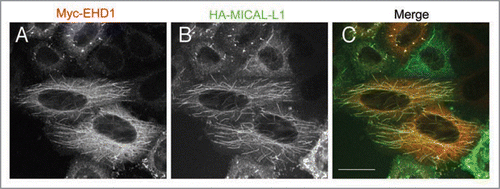
The interaction of MICAL-L1 and EHD1 was mediated exclusively by the first of the two asparagine-proline-phenylalanine (NPF) motifs, as mutation of the residues comprising this motif to alanine completely abrogated the binding between the two proteins. We analyzed the localization of MICAL-L1 using specific antibodies and found that it primarily associated with the tubular membranes of the ERC, which highly co-localized with EHD1-positive tubules (see ). Surprisingly, MICAL-L1 continued to associate with tubules even in cells depleted of EHD1, again suggesting that EHD1 does not generate the tubules but localizes to pre-existing tubular membranes. In support of this notion, we also found that the MICAL-L1 NPF motifs were not required for its own tubular localization, but rather the C-terminal coiled-coil region was both essential and sufficient for this subcellular distribution.Citation13
Since the association of MICAL-L1 with tubules was independent of EHD1 expression, we next analyzed if the reverse was true; if MICAL-L1 was required for EHD1 association with tubules. Interestingly, in cells lacking MICAL-L1, there was a very dramatic and almost complete loss of EHD1 localization to tubules, indicating that MICAL-L1 is essential for the association of EHD1 with tubular membranes, and thus for its function in endocytic recycling. Previous studies had reported that MICAL-L1 directly binds to GTP-bound Rab8a,Citation16,Citation17 a Rab protein involved in endocytic recyclingCitation18 that colocalizes with EHD1 positive tubular endosomes.Citation12 To test if MICAL-L1 was the critical link between these two recycling regulators, we performed MICAL-L1 knockdown and analyzed the distribution and association of EHD1 and Rab8a with the tubules. MICAL-L1 was found to regulate the recruitment of both EHD1 and Rab8a on to the tubular membranes and was responsible for their co-association on these tubules. On the other hand, no effect on localization of MICAL-L1 or EHD1 was observed upon Rab8a knockdown. Finally, MICAL-L1 depletion impaired the recycling of both transferrin and β1 integrin receptors and caused their accumulation in the ERC indicating that MICAL-L1 regulates receptor recycling from the ERC back to the plasma membrane.Citation13
In conclusion, we have identified MICAL-L1 as a novel interaction partner for EHD1 that may link EHD1 and Rab8a on tubular recycling endosomes. Moreover, MICAL-L1 appears to be an unusual type of Rab effector that is recruited to membranes independently of Rab8a expression and is itself involved in regulating membrane association of Rab8a. Future studies are needed to address whether other Rab proteins recruit MICAL-L1 to tubular membranes and to elucidate the mechanism(s) by which the C-terminal coiled-coil regions of MICAL-L1 associates with tubular membranes, thereby regulating the recruitment of EHD1 and Rab8a to these membranes.
Figures and Tables
Figure 1 Domain Architecture of MICAL-family of proteins. FAD: Flavin-adenine dinucleotide binding domain, CH: Calponin Homology domain, LIM: LIM domain, NPF: Asn-Pro-Phe motif, CC: Coiled coil domain.
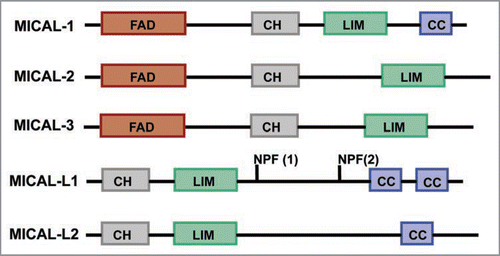
Figure 2 Co-localization of EHD1 and MICAL-L1 to tubular recycling endosomes. HeLa cells were co-transfected with Myc-tagged EHD1 (A) and HA-tagged MICAL-L1 (B). After 16 h, cells were fixed/permeabilized and incubated with anti-Myc, and anti-HA primary antibodies, and appropriate secondary antibodies before being mounted on cover-slides. Images were obtained from a Zeiss LSM 5 confocal microscope, using a 63x objective with a numerical aperture of 1.4. Bar, 10 µm.
Acknowledgements
We gratefully acknowledge the support of NIH grants GM074876 (S.C.) and P20 RR018759 (N.N. and S.C.), and the American Heart Association (M.S.).
Addendum to:
References
- Hopkins CR. Intracellular routing of transferrin and transferrin receptors in epidermoid carcinoma A431 cells. Cell 1983; 35:321 - 330
- Maxfield FR, McGraw TE. Endocytic recycling. Nat Rev Mol Cell Biol 2004; 5:121 - 132
- Ullrich O, et al. Rab11 regulates recycling through the pericentriolar recycling endosome. J Cell Biol 1996; 135:913 - 924
- Grant BD, Caplan S. Mechanisms of EHD/RME-1 Protein Function in Endocytic Transport Traffic 2008
- Caplan S, et al. A tubular EHD1-containing compartment involved in the recycling of major histocompatibility complex class I molecules to the plasma membrane. EMBO J 2002; 21:2557 - 2567
- Grant B, et al. Evidence that RME-1, a conserved C. elegans EH-domain protein, functions in endocytic recycling. Nat Cell Biol 2001; 3:573 - 579
- Guilherme A, Soriano NA, Furcinitti PS, Czech MP. Role of EHD1 and EHBP1 in perinuclear sorting and insulin-regulated GLUT4 recycling in 3T3-L1 adipocytes. J Biol Chem 2004; 279:40062 - 40075
- Jovic M, et al. EHD1 regulates beta1 integrin endosomal transport: effects on focal adhesions, cell spreading and migration. J Cell Sci 2007; 120:802 - 814
- Lin SX, Grant B, Hirsh D, Maxfield FR. Rme-1 regulates the distribution and function of the endocytic recycling compartment in mammalian cells. Nat Cell Biol 2001; 3:567 - 572
- Jovic M, et al. Eps15 Homology Domain 1-associated Tubules Contain Phosphatidylinositol-4-Phosphate and Phosphatidylinositol-(4,5)-Bisphosphate and Are Required for Efficient Recycling. Mol Biol Cell 2009; 20:2731 - 2743
- Daumke O, et al. Architectural and mechanistic insights into an EHD ATPase involved in membrane remodelling. Nature 2007; 449:923 - 927
- Roland JT, et al. Myosin Vb interacts with Rab8a on a tubular network containing EHD1 and EHD3. Mol Biol Cell 2007; 18:2828 - 2837
- Sharma M, et al. MICAL-L1 Links EHD1 to Tubular Recycling Endosomes and Regulates Receptor Recycling. Mol Biol Cell 2009;
- Suzuki T, et al. MICAL, a novel CasL interacting molecule, associates with vimentin. J Biol Chem 2002; 277:14933 - 14941
- Terman JR, et al. MICALs, a family of conserved flavoprotein oxidoreductases, function in plexinmediated axonal repulsion. Cell 2002; 109:887 - 900
- Fukuda M, Kanno E, Ishibashi K, Itoh T. Large scale screening for novel rab effectors reveals unexpected broad Rab binding specificity. Mol Cell Proteomics 2008; 7:1031 - 1042
- Yamamura R, et al. The interaction of JRAB/ MICAL-L2 with Rab8 and Rab13 coordinates the assembly of tight junctions and adherens junctions. Mol Biol Cell 2008; 19:971 - 983
- Hattula K, et al. Characterization of the Rab8- specific membrane traffic route linked to protrusion formation. J Cell Sci 2006; 119:4866 - 4877