Abstract
Yeast colonies growing on solid medium begin at a particular point in their development to produce volatile ammonia and to alkalize their surroundings.1 Ammonia serves as a long-range signal between neighboring colonies and was shown to influence various aspects of colony biology including metabolic reprogramming and differentiation. In a recent paper we presented the impact of deleting of key stress defense enzymes on ammonia signaling and colony development. New findings suggest that it is not stress defense, but rather proper development guided by ammonia signaling and related metabolic changes that are important factors in the long-term survival of a colony cell population.
Yeast colonies growing on solid medium begin at a particular point in their development to produce volatile ammonia and to alkalize their surroundings. Citation1 Ammonia serves as a long-range signal between neighboring colonies and was shown to influence various aspects of colony biology including metabolic reprogramming and differentiation. In a recent paper we presented the impact of deleting of key stress defense enzymes on ammonia signaling and colony development. New findings suggest that it is not stress defense, but rather proper development guided by ammonia signaling and related metabolic changes that are important factors in the long-term survival of a colony cell population.
Ammonia signaling was found to be connected with extensive expression changesCitation2 including metabolic reprogramming consisting of the repression of mitochondrial oxidative phosphorylation and the mitochondrial tricarboxylic acid cycle alongside the activation of amino acid and nucleotide metabolism and some poorly understood branches of carbon metabolism (e.g., genes ICL2, CIT3, FDH1) and transport (e.g., genes ATO1, ATO2, ATO3, JEN1). In parallel, the expression of stress-related genes, including those for oxidative stress defense enzymes, falls during colony development. This is rather surprising, since in liquid cultures aging results in stress factor accumulation, and the culture fitness is proportional to stress defense enzyme activities. What is then the importance of stress defense in colonies?
Studies of the differentiation between central and margin region cell populations of the colony revealed that cells with apoptotic-like features (e.g., reactive oxygen species (ROS) production, chromatin fragmentation, half-empty shrunken cells) are restricted to the colony centre, leaving colony margin cells healthy and fully able to propagate the colony towards an uncolonized area.Citation3 Similar centre-margin differences were found in the expression of various proteins and antioxidant enzyme activities.Citation4 Hence, there is clear evidence that cells located in distinct colony regions differentiate and that the starting point of this differentiation approximately correlates with the initiation of ammonia production. This raises the question as to whether differentiation is connected to ammonia signaling.
Additional results came from studies of colonies with a deleted SOK2 gene, which encodes for a transcription regulator involved in various physiological processes. Colonies of this sok2Δ strain were unable to produce ammonia or activate variety of metabolic genes typical for ammonia-connected metabolic reprogramming.Citation5 Even more interesting was the fact that these colonies also have an impaired colony differentiation.Citation3 Unlike in the wild type, there was only a slight difference between the sok2Δ colony central and margin cells in the studied parameters. Is this lack of centre-margin differentiation of sok2Δ colonies connected to the reduction in ammonia signaling and inability to undergo metabolic changes?
To answer these questions, we performed a detailed analysis of colonies formed by strains deleted in key enzymes of oxidative stress defense: i.e., sod1Δ deficient in cytosolic superoxide dismutase, sod2Δ lacking mitochondrial superoxide dismutase and ctt1Δ with the deletion of the gene for cytosolic catalase. As for their ability to produce ammonia and undergo ammonia related metabolic changes, sod1Δ was fully able to produce ammonia and activate genes typical for the ammonia- connected metabolic switch (e.g., CIT3, ATO1, ATO3, JEN1), while the sod2Δ strain was incapable of both ammonia production and sufficient induction of metabolic genes. Preliminary microarray data for the sod2Δ strain also revealed other similarities between sod2Δ and sok2Δ colonies at the transcriptional level, especially the lack of induction of amino acid, nucleotide and other metabolic changes typical for the ammonia-producing phase of wt colonies (unpublished data). Thus, we had an experimental model where the deletion of a single enzymatic activity located in different compartments (cytosol vs. mitochondria) led to differing effects on ammonia signaling and the connected alternative metabolism activation. We then examined in detail the differentiation in several stress-related physiological parameters (ROS production, cell death occurrence, thermoresistance, activity of oxidative stress defense enzymes) between central and margin regions in colonies formed by oxidative-stress-impaired mutants. It turned out that center-margin differentiation is dependent on the ability of the particular strain to undergo the ammonia-mediated metabolic switch. Hence, in sod2Δ colonies we observed a similar consequence of the absence of ammonia signaling as in sok2Δ colonies: an inhibition of the activation of the metabolic changes and consequently an inhibition of sod2Δ colony differentiation. This complex colony phenotype was thus found in strains lacking two functionally unrelated proteins, the pleiotropic transcription regulator Sok2p and ROS scavenging enzyme Sod2p, indicating the key role of ammonia-guided metabolic and possibly other changes. This finding is also supported by observations of ctt1Δ colonies, which produce a reduced amount of ammonia and, correspondingly, exhibit a level of activation of metabolic changes as well as differentiation that is in-between the wt and sod2Δ. Interestingly, the overall level of ROS (superoxide or hydrogen peroxide) was not elevated in aging colonies of any of the stress-defense mutants compared to the wt colonies.
The above results raise the possibility that Sod2p and/or cellular ROS homeostasis are somehow involved in signaling pathways leading to ammonia production and the subsequent metabolic switch. This hypothesis predicts that a higher mitochondrial superoxide level in the early stages of colony development could somehow impair the onset of ammonia signaling. And indeed, in contrast to aging colonies, superoxide production is elevated in the mitochondria of very young sod2Δ colonies (1- and 3-day old colonies, unpublished data). One could imagine several mechanisms for the ROS imbalance effect, one of them being that mitochondrial ROS could block or crosstalk with pathways leading to ammonia signaling. Several mitochondrial redox-sensing pathways were proposed in higher organisms that regulate either mitochondrial physiology or nuclear gene expressionCitation6–Citation8 but none in yeast so far. Alternatively, mitochondrial ROS may act directly by damaging particular enzyme(s) necessary for ammonia generation. A number of amino acid metabolic genes are concentrated in mitochondria, many of which are induced during the ammonia-production phase. Since it has been shown that amino acid metabolism is source of ammonia,Citation9 oxidative damage to some enzymes could affect ammonia production at the enzymatic level. Finally, sod2Δ could have another role in cellular signaling besides its enzymatic activity. Mitochondrial superoxide dismutase was recently suggested to have a regulatory role in DNA repair machinery and chromatin remodeling in yeast.Citation10
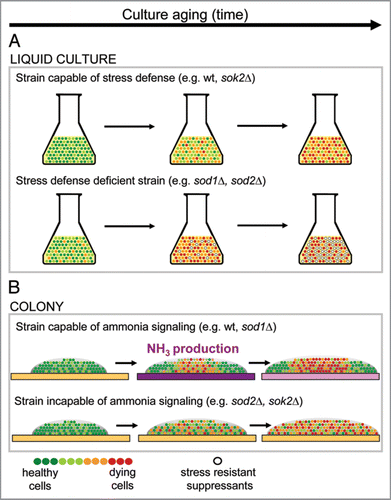
Stress-defense-deficient mutants, especially sod1Δ and sod2Δ, are severely damaged in liquid cultivations, as documented by their slow growth, high ROS level and rapid cell dying.Citation11–Citation13 In colonies, however, the situation is different. sod2Δ and ctt1Δ colonies (incapable of ammonia signaling) lack differentiation, which results in a large proportion of stressed and dead cells at the colony margin. In contrast, sod1Δ, the strain most damaged in liquid cultivations, is as prosperous as the wt under colonial conditions. Since the sod1Δ cell population is known to efficiently suppress defects evoked by oxidative stress by accumulating stress-resistant suppressor mutations, it was a surprise that no such mutants were detected in sod1Δ colonies. sod1Δ cells taken from colonies, however, acquired some new heritably stable physiological features, such as different growth properties and survival rate in liquid cultures, showing that this strain, although developing in a colonial environment, is prone to changes (mutations or epigenetic switches). The lack of accumulation of stress-resistant suppressor mutants thus indicates that in colonies there is no selection pressure in favor of stress-resistant mutations. This demonstrates that completely different factors are important for survival in homogeneous, aerated, fast growing liquid shaken cultivations and in the differentiated multicellular population of a colony (). A colony represents a highly complex and heterogeneous population developing in an environment where gradients of various compounds, nutrients or gases form many different niches, where each cell occupies its particular space and cell-to-cell interactions are preserved. In this respect, the colonial model resembles to some extent the tissues of real multicellular organisms and could contribute to uncovering and understanding new aging and stress-related processes in slowly dividing or non-dividing differentiated cell populations.
Figures and Tables
Figure 1 Different factors are important for survival in liquid cultures and in colonies. (A) Survival in liquid culture is dependent on stress defense. Stress-defense-deficient population dies rapidly unless stress-resistant mutants appear and overgrow the dying population. (B) Survival in a colony is dependent on metabolic adaptation of the population. Colony cell population switches metabolism as a result of ammonia signal which enables survival of cells at colony margin. Cells in colony centre die and provide nutrients for the benefit of cells at the colony margin. Development of a colony incapable of ammonia signaling is not orchestrated, cells are not able to activate their alternative metabolism and consequently cell death is spread throughout the whole colony. Color of the agar-medium indicates pH changes during colony development: yellow, pH below 6.5; violet, pH above 6.5.
Acknowledgements
This work was supported by Grant Agency of the Czech Republic (204/08/0718), by Ministry of Education (LC531, MSM0021620858 and AV0Z50200510) and by the Howard Hughes Medical Institute (International Research Award 55005623 to Z.P.).
Addendum to:
References
- Palkova Z, Janderova B, Gabriel J, Zikanova B, Pospisek M, Forstova J. Ammonia mediates communication between yeast colonies. Nature 1997; 390:532 - 536
- Palkova Z, Devaux F, Ricicova M, Minarikova L, Le Crom S, Jacq C. Ammonia pulses and metabolic oscillations guide yeast colony development. Mol Biol Cell 2002; 13:3901 - 3914
- Vachova L, Palkova Z. Physiological regulation of yeast cell death in multicellular colonies is triggered by ammonia. J Cell Biol 2005; 169:711 - 717
- Vachova L, Kucerova H, Devaux F, Ulehlova M, Palkova Z. Metabolic diversification of cells during the development of yeast colonies. Environ Microbiol 2009; 11:494 - 504
- Vachova L, Devaux F, Kucerova H, Ricicova M, Jacq C, Palkova Z. Sok2p transcription factor is involved in adaptive program relevant for long term survival of Saccharomyces cerevisiae colonies. J Biol Chem 2004; 279:37973 - 37981
- Balmer Y, Vensel WH, Tanaka CK, Hurkman WJ, Gelhaye E, Rouhier N, et al. Thioredoxin links redox to the regulation of fundamental processes of plant mitochondria. Proc Natl Acad Sci USA 2004; 101:2642 - 2647
- Storz P. Mitochondrial ROS-radical detoxification, mediated by protein kinase D. Trends Cell Biol 2007; 17:13 - 18
- Armstrong JS, Whiteman M, Yang H, Jones DP. The redox regulation of intermediary metabolism by a superoxide-aconitase rheostat. Bioessays 2004; 26:894 - 900
- Zikanova B, Kuthan M, Ricicova M, Forstova J, Palkova Z. Amino acids control ammonia pulses in yeast colonies. Biochem Biophys Res Commun 2002; 294:962 - 967
- Bonatto D. A systems biology analysis of proteinprotein interactions between yeast superoxide dismutases and DNA repair pathways. Free Radic Biol Med 2007; 43:557 - 567
- Davidson JF, Whyte B, Bissinger PH, Schiestl RH. Oxidative stress is involved in heat-induced cell death in Saccharomyces cerevisiae. Proc Natl Acad Sci USA 1996; 93:5116 - 5121
- Longo VD, Gralla EB, Valentine JS. Superoxide dismutase activity is essential for stationary phase survival in Saccharomyces cerevisiae. Mitochondrial production of toxic oxygen species in vivo. J Biol Chem 1996; 271:12275 - 12280
- Gralla EB, Valentine JS. Null mutants of Saccharomyces cerevisiae Cu,Zn superoxide dismutase: characterization and spontaneous mutation rates. J Bacteriol 1991; 173:5918 - 5920