Abstract
A single and purified clone of foot-and-mouth disease virus diversified in cell culture into two subpopulations that were genetically distinct. The subpopulation with higher virulence was a minority and was suppressed by the dominant but less virulent one. These two populations follow the competition-colonization dynamics described in ecology. Virulent viruses can be regarded as colonizers because they killed the cells faster and they spread faster. The attenuated subpopulation resembles competitors because of its higher replication efficiency in coinfected cells. Our results suggest a new model for the evolution of virulence which is based on interactions between components of the quasispecies. Competition between viral mutants takes place at two levels, intracellular competition, and competition for new cells. The two strategies are subjected to density-dependent selection.
Viral populations are dynamic ensembles of genetically diverse genomes, referred to as viral quasispecies, that arise from of error-prone genome replication catalyzed by low fidelity RNA-dependent RNA or DNA polymerases. Genomes in the population are subjected to continuous competition, and both, individual replicative fitness and interactions among viruses within the population determine which variants will increase their relative abundance and which will remain minority components of the quasispecies ().Citation1 RNA viruses are thus endowed with great adaptability to changing environments, and even single clones have the potential to rapidly generate explosive diversity after few rounds of replication. Studies with viral mutants that copy their genetic material with increased fidelity have documented that high mutation rates are essential for adaptation of the virus to a complex biological environment.Citation2,Citation3 In the case of our recent study with the important animal pathogen foot-and-mouth disease virus (FMDV), a single purified clone evolved during serial cell culture infections to give rise to two phylogenetically different progeny subpopulations. Remarkably, the subpopulation with higher fitness (replication capacity) was found to be the minority in the quasispecies.Citation4
Two interesting questions are raised by this result: (1) Which selective pressure favoured the divergence of a single purified clone into two defined evolutionary trajectories? The question is particularly intriguing if we consider that an established cell line is the most homogeneous environment available under laboratory conditions to support sustained viral replication. (2) Why did the high-fitness virus remain at low frequency over an extended course of serial infections?
A detailed phenotypic analysis of the two subpopulations revealed striking differences between them. The minority high-fitness subpopulation manifested a high-virulence phenotype, measured as the cell killing rate. The dominant subpopulation showed higher efficiency in progeny production in coinfected cells, and interfered with replication of the virulent variants resulting in a delay of cell killing. The infections were carried out at high multiplicity of infection (MOI), i.e., at a high density of viruses relative to the number of cells. High-MOI infections allow the occurrence of multiple coinfections and interaction between variants within coinfected cells, thus explaining the dominance of the low-fitness subpopulation.
The two phenotypes characterized within the viral population resemble the evolutionary strategies of competition and colonization defined in ecology.Citation5 Virulent viruses can be regarded as colonizers, because they kill the cell faster which allows them to spread faster. In turn, the interfering viruses can be regarded as competitors, because they are more efficient in exploiting the local resources which in this case are provided by individual cells.
Competition-colonization dynamics are typically observed in spatially structured habitats. In the case of viral infections, any set of cells can be regarded as a patchy habitat for viral replication, where each cell constitutes a patch (). The patchiness of the cell culture, i.e., the fact that the culture is divided into individual cells, provides two basic ecological niches for viral replication, namely competition for new patches (uninfected cells), and local competition inside a single resource (coinfected cells). In our experiments, these two niches directed the diversification of the progeny of a single and clonal sequence with consequences for the virulence phenotype of the population as a whole.
Fitness and virulence are not necessarily directly correlated traits in RNA viruses.Citation6,Citation7 Moreover, the evolution of virulence in viruses has typically been addressed applying models that either did not take into account coinfection of cells at all, or that assumed virulent viruses to be more efficient during coinfections.Citation8 Consequently, many of these models predict an increase in the average virulence of the population. Based on our experimental results, we developed a new mathematical approach based on the SIR model that describes the dynamics of susceptible and infected cells and the production of the two viral subpopulations as a microepidemic in cell culture.Citation9 Specifically, the model considered the existence of coinfetcted cells (). Analysis of the model revealed that the coinfection dynamics depends only on the two parameters that best describe the competition and colonization phenotypes, namely the difference in virulence between the two viruses and their intracellular competitiveness. Model simulations also predicted a density-dependent outcome of the coinfection dynamics. Under conditions of low MOI, virulent colonizers had an advantage because they spread faster through the unoccupied space of susceptible cells. In contrast, under high-density conditions, coinfections were more frequent and the competition strategy had higher success. Our new mathematical approach therefore establishes density-dependent selection as a link between fitness and virulence. The model is a promising tool to study the evolution of virulence, offering a broader scenario in which viral populations have the potential to evolve either towards increased virulence or towards attenuation.
Density-dependent selection of viruses and suppression of high-fitness clones has been described in other viral systems,Citation10–Citation14 which strongly suggests that competitioncolonization dynamics could be a general evolutionary mechanism for viruses. During coinfection, different members of the quasispecies can interact between them in the form of negative dominant mutants, also referred to as interfering viruses.Citation10,Citation15–Citation19
A growing body of evidence indicates that mutant spectra of RNA virus quasispecies are ensembles of interacting mutants rather than mere collections of mutants ranked according to individual fitness and mutation rate as in the classic mutation-selection equilibrium. Fitness is a collective property of the virus population as also recognized for a number of cellular consortia.Citation20 Evidence for RNA viruses includes the presence of memory genome subpopulations which can alter the evolutionary outcome of the ensemble, complementation and interference among components of the mutant spectra that were reproduced with reconstructed quasispecies, pathogenic potential of viruses or response to antiviral treatments which are influenced by the amplitude of the mutant spectrum and by its composition.Citation1,Citation15,Citation21,Citation22 The interplay between inhibitor-escape mutants and defector genomes produced by mutagenesis underlies a possible advantage of a sequential versus combination antiviral therapy.Citation23 In our recent study,Citation4 a link between quasispecies behavior and ecology can explain modulation of virulence. This phenomenon was mediated by high mutation rates and the potential of the viral system to select a differentiation trajectory for the benefit of the ensemble.
Figures and Tables
Figure 1 Quasispecies replication dynamics of RN A viruses. The central scheme represents a characteristic heterogeneous distribution of genomes. Each line represents a genome with mutations depicted as symbols. Average consensus sequence is depicted below the mutant spectrum. The central initial population can evolve towards increased fitness (on the right) when replication takes place with high population size. When the population undergoes bottlenecks (on the left) the fitness of the population drops by the action of Müller’s ratchet. Virulence evolution follows an independent trajectory from the one observed for relative fitness; low fitness viruses can have a high virulence phenotype.
Figure 2 Continuous versus discrete resources. (A) Two bacterial populations growing independently on the top of an agar layer. The agar can be considered a continuous resource. (B) Two viruses (colonizers in green and competitors in blue) infect initially a cell culture. Colonizers complete faster the infection cycle and the released progeny spreads through the unoccupied, susceptible cells. (C) Under high density of viruses almost all cells are coinfected and interaction between viruses takes place inside the single resource (the infected cell); cells can thus be considered a discrete (patchy) resource.
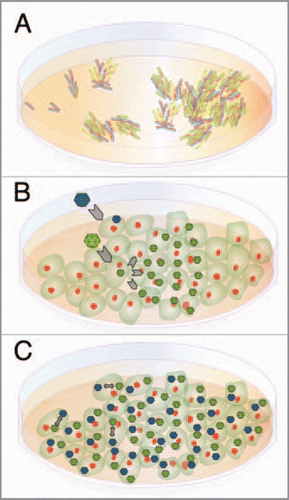
Figure 3 Competition-colonization trade-off in a two-virus system. (A) The scheme represents a two virus competition (blue, competitor; green, colonizer). Virulence, ai, is defined as cell killing rate. Competitiveness, c, is the proportion of each virus produced in a coinfected cell. (B) The plot illustrates the model prediction of a density-dependent outcome of the coevolutionary dynamics of a colonizer and a competitor in cell culture. Shown is the critical intracellular competitveness, c, defined as the fraction of colonizer offspring that a coinfected cell will produce, as a function of the ratio of the colonizer virulence to the competitor virulence, a, for different initial viral densities. For any given initial viral density, colonizers are predicted to win the competition if the viral phenotypes (a, c) fall below the respective curve. Otherwise, competitors are predicted to outcompete colonizers. For further details see reference 4.
Acknowledgements
Work at CBMSO supported by grant BFU2008-02816/BMC and Fundación Ramón Areces. CIBERehd (Centro de Investigación Biomédica en Red de Enfermedades Hepáticas y Digestivas) is funded by Instituto de Salud Carlos III.
Addendum to:
References
- Domingo E. Knipe DM, Howley PM. Virus Evolution. Fields Virology 2007; 5th ed. Philadelphia Lappincott Williams & Wilkins 389 - 421
- Pfeiffer JK, Kirkegaard K. Increased fidelity reduces poliovirus fitness under selective pressure in mice. PLoS Pathog 2005; 1:102 - 110
- Vignuzzi M, Stone JK, Arnold JJ, Cameron CE, Andino R. Quasispecies diversity determines pathogenesis through cooperative interactions in a viral population. Nature 2006; 439:344 - 348
- Ojosnegros S, Beerenwinkel N, Antal T, Nowak MA, Escarmis C, et al. Competition-colonization dynamics in an RNA virus. Proc Natl Acad Sci USA 2010; 107:2108 - 2112
- Tilman D, May RM, Lehman CL, Nowak MA. Habitat destruction and the extinction debt. Nature 1994; 371:65 - 66
- Herrera M, Garcia-Arriaza J, Pariente N, Escarmis C, Domingo E. Molecular basis for a lack of correlation between viral fitness and cell killing capacity. PLoS Pathog 2007; 3:53
- Carrasco P, de la Iglesia F, Elena SF. Distribution of fitness and virulence effects caused by singlenucleotide substitutions in Tobacco Etch virus. J Virol 2007; 81:12979 - 12984
- May RM, Nowak MA. Coinfection and the evolution of parasite virulence. Proc Biol Sci 1995; 261:209 - 215
- Nowak MA, May RM. Virus Dynamics. Mathematical Principles of Immunology and Virology 2000; New York Oxford University Press Inc
- de la Torre JC, Holland JJ. RNA virus quasispecies populations can suppress vastly superior mutant progeny. J Virol 1990; 64:6278 - 6281
- Novella IS, Reissig DD, Wilke CO. Densitydependent selection in vesicular stomatitis virus. J Virol 2004; 78:5799 - 5804
- Sevilla N, Ruiz-Jarabo CM, Gomez-Mariano G, Baranowski E, Domingo E. An RNA virus can adapt to the multiplicity of infection. J Gen Virol 1998; 79:2971 - 2980
- Turner PE, Chao L. Prisoner’s dilemma in an RNA virus. Nature 1999; 398:441 - 443
- Bull JJ, Millstein J, Orcutt J, Wichman HA. Evolutionary feedback mediated through population density, illustrated with viruses in chemostats. Am Nat 2006; 167:39 - 51
- Crowder S, Kirkegaard K. Trans-dominant inhibition of RNA viral replication can slow growth of drug-resistant viruses. Nature Genet 2005; 37:701 - 709
- Perales C, Mateo R, Mateu MG, Domingo E. Insights into RNA virus mutant spectrum and lethal mutagenesis events: replicative interference and complementation by multiple point mutants. J Mol Biol 2007; 369:985 - 1000
- Baulcombe DC. Mechanisms of pathogen-derived resistance to viruses in transgenic plants. Plant Cell 1996; 8:1833 - 1844
- Grande-Perez A, Lazaro E, Lowenstein P, Domingo E, Manrubia SC. Suppression of viral infectivity through lethal defection. Proc Natl Acad Sci USA 2005; 102:4448 - 4452
- Holland JJ. Fields BM, Knipe DM. Defective viral genomes. Virology 1990; 151-65 New York Raven Press
- Mas A, López-Galíndez C, Cacho I, Gómez J. Unfinished stories on viral quasispecies and Darwinian views of evolution. J Mol Biol 2010; 397:865 - 877
- Perales C, Martin V, Ruiz-Jarabo CM, Domingo E. Monitoring sequence space as a test for the target of selection in viruses. J Mol Biol 2005; 345:451 - 459
- Grande-Perez A, Gomez-Mariano G, Lowenstein PR, Domingo E. Mutagenesis-induced, large fitness variations with an invariant arenavirus consensus genomic nucleotide sequence. J Virol 2005; 79:10451 - 10459
- Perales C, Agudo R, Tejero H, Manrubia SC, Domingo E. Potential benefits of sequential inhibitor-mutagen treatments of RNA virus infections. PLoS Pathog 2009; 5:1000658