Abstract
Single-molecule research is emerging as one of the fastest growing fields within the biosciences. Historically, most of the techniques employed have operated largely in the world of the test tube in which the components of the biological system under investigation have been extracted and purified from cells to reduce them to just the key ingredients under study, and this research has involved novel, pioneering methods of biophysics to obtain single-molecule measurements. What has emerged recently is the technical ability to now perform key single-molecule experiments whilst retaining the native biological context – namely to do single-molecule experiments on functional living cells. This presents essentially a new science of “single-molecule cell biology”, which combines classical cell biology approaches with modern single-molecule biophysics. Here, key recent studies which have pushed back the boundaries of this field are discussed.
Conventional experimental approaches to date have utilized so-called “bulk ensemble-average” techniques, namely to measure biological properties by in effect taking an average measurement resulting from perhaps several thousand individual molecular interactions within a given biological system. However, many important processes in biology occur at the level of one or only a few single molecules, since the copy-number of the relevant molecules is often relatively low in each given cell. A more revealing experimental approach therefore is to utilize powerful single-molecule experimental techniques to the study of complex biological systems. If one is able to observe the properties of relatively few individual molecular interactions one by one then we can discover not only how the normal ensemble-average behavior can be generated, but also witness the unpredictable behavior normally masked by the usual ensemble-averaging process, namely the actual, individual, causal molecular events that give rise ultimately to collective properties; in essence taking a “bottom up” molecular-level approach. At present we have a largely static picture for the structure of molecules and molecular complexes in living cells, which tells us nothing about the dynamics and how they operate in cells. Single-molecule methods are needed to reveal these details.
Many of the existing techniques of conventional single-molecule bioscience involve fundamentally in vitro assays. Namely, the use of experimental techniques for which the contents of cells under study have been removed from their original biological context. Although many of these experiments are ingenious and highly controlled they still present a potentially serious issue in the correct interpretation of the relevance of such data from a cell-level perspective—there is a serious danger of throwing out the baby with the bathwater. It is clear that layers of complex feedback exist between biological components at many different time and length scales, and furthermore that biological functionality in the living cell is generally localized in a highly specific manner to distinct sub-cellular regions. The key thus is to design challenging complementary experiments, using both purified test tube level experiments to give us information on specific key components whilst, with the requisite technology, monitoring processes in other related experiments from a whole cell perspective but at a single-molecule precise level with the original biological context intact. While it is clear that a single protein molecule must function within the context of an entire cell, it has only recently become technologically feasible to obtain data at this functional level. With the help of state-of-the-art genetics methods, individual protein molecules can now be observed within a living, functioning cell and their exchange with other protein molecules monitored in several examples of complex, functioning biological systems.Citation1 These utilize predominantly advanced fluorescence microscopy since such approaches permit minimal perturbation to the biological system under study whilst offering the contrast required to image at a single-molecule level.
Measuring Gene Expression
A recent study investigated the central dogma of molecular biology,Citation2 namely that the DNA genetic code is first transcribed into mRNA, which is ultimately translated into protein molecules. Here, the authors used the model system of the lac operonCitation3 in the bacterium Escherichia coli to observe bursts of gene expression activity. They were able to encode a membrane protein Tsr fused to yellow fluorescent protein (YFP) into the genome but under control of the lac promoter to act as a reporter for gene expression from that operon. This particular Tsr protein was selected for two reasons. Firstly, the native Tsr with no fluorescent tag attached is expressed as part of a membrane receptor complex at levels of several thousand per cell. Thus, the relatively meagre expression bursts stemming from the lac operon would most likely not perturb this native physiology. Secondly, since Tsr is integrated into the cell membrane its mobility will be typically a thousand times lower than a protein of similar molecular weight expressed in the cytoplasm. This is important since it meant that relatively slow imaging at 100 ms per frame using a conventional epifluorescence microscope was fast enough to image to single Tsr protein molecules unblurred.
By monitoring fluorescence images in single living E. coli cells over typically eight cell division generations the authors built up a picture for the activity of gene expression bursts. Using the intensity of diffraction-limited fluorescence spots of width ∼300 nm associated with YFP they could estimate how many molecules of Tsr protein were expressed in every burst of activity, and also correlate this to the number of mRNA molecules associated with each burst. They concluded that a single molecule of mRNA was associated with each gene expression burst, but that the number of protein molecules translated from this was variable, averaging 4–5 to every one mRNA. This is consistent with a scheme in which transcription of a single mRNA molecule by one or more RNA polymerase enzyme complexes can result in generating multiple copies of a protein molecule before the mRNA is degraded marking the end of that particular burst of gene expression activity.
Counting the Dynamic Protein Components of a Molecular Motor
In an unrelated study, E. coli was again employed, but in monitoring the activity of functional molecular complexes.Citation4 Here, a green fluorescent protein (GFP) tag was fused to a protein MotB implicated in the generation of torque in the bacterial flagellar motor, this time under control of the native MotB promoter and equivalent to wild-type levels of expression. The flagellar motor is a remarkable biological nano-machine which ultimately drives the swimming of a bacterial cell.Citation5 It is generated from a coordinated expression of ∼50 genes resulting in a membrane-spanning structure which can rotate an external filament of up to ∼10 micrometers in length hundreds of times per second. The motor is ∼50 nm in diameter and thought to be composed of ∼13 different proteins. It is fuelled by an ion flux of either protons or sodium ions, depending upon the species, across the cell membrane and has all the components one might expect from a rotary motor in the macro world, such as “stator” units that push an axial “rotor” around.
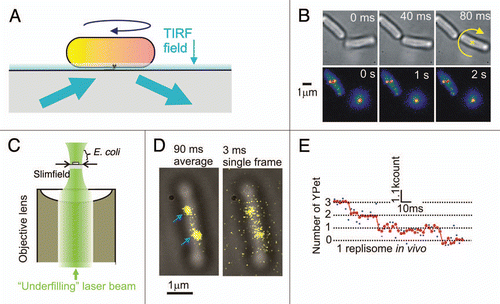
By tethering a single living E. coli cell via one of its filaments to a microscope coverslip the authors could observe the cell rotating about that point (). This indicated that not only was the cell alive but also that the molecular complex of the flagellar motor was functional. Using total-internal-reflection fluorescence (TIRF) microscopy (), they were able to increase the imaging contrast to allow single ∼300 nm diameter diffraction-limited spots of fluorescence intensity to be visualized, consistent with the size and location of the flagellar motor. Using a phenomenon of stepwise photobleaching, namely that dye molecules such as GFP will photobleach stochastically to a dark state in a step-like manner, the authors could count how many of the GFP molecules were associated with each spot, and then estimate the stoichiometry of the MotB protein within a functional motor. This indicated a broad distribution centred on ∼22 molecules, which further indicated that there were ∼11 individual stator units per motor since two MotB molecules were known to be used in each stator.
Using a combination of fluorescence recovery after photobleaching (FRAP) and fluorescence loss in photobleaching (FLIP) the authors discovered that the MotB protein diffused in the membrane when it was not integrated into a motor, but also that MotB molecules were not permanent features but rather turned over by some unresolved exchange mechanism with the membrane pool on average once every ∼30 seconds.
A further study from the same team on the same motor complex suggested that in some instances molecular turnover may be dependent upon a signalling factor. Here they tagged a protein called FliM with the fluorescent protein YPet.Citation6 FliM is used in the bacterial flagellar motor as part of a switching complex which causes the motor to either stop or change direction, depending on the species. The investigators used a similar stepwise photobleaching approach to measure the stoichiometry of FliM, this time indicating more like ∼30 molecules per motor, and also assessed molecular turnover using FRAP and FLIP on a variety of mutated cell strains in which the mutations affected the activity or presence of a response regulator protein, CheY, known to be involved in signalling between receptors on the membrane surface and the rotational state of the motor. This indicated FliM also turned over with a similar time scale to MotB, but here the turnover relied upon the presence of the phosphorylated, activated form of the CheY molecule.
Probing DNA Replication
In another recent study, investigators again used the model E. coli bacterium to explore the architecture behind DNA replication.Citation7 The multiprotein complex called the replisome that replicates DNA had been studied extensively previously in vitro, but its true composition and architecture in the living cell was essentially unknown. Replisomes are dynamic, complex machines that replicate DNA by copying a template from the leading-strand continuously whilst copying that from the lagging-strand template in a discontinuous fashion. Typically, the replisome combines the activities of at least 11 proteins during genome replication in a coordinated fashion.Citation8,Citation9 Here, the authors constructed 10 different cell strains, each of which contained YPet fused to a protein known to be a component of the replisome. The YPet DNA was inserted under control of the native promoter of the replisome protein under study and so was expressed at normal physiological levels. However, since these proteins are expressed in the cytoplasm their mobility is significantly greater compared to the higher viscosity environment of membrane-integrated proteins, and so conventional “video-rate” fluorescence microscopy (typically sampling at a speed of tens of milliseconds per image frame) was too slow to image the proteins unblurred. To overcome this problem, the authors used a method of fluorescence illumination called slimfieldCitation10 which concentrated the laser excitation light into a much smaller area over the cell sample (). This resulted in a much greater intensity of excitation sufficient to overcome camera noise when lowering the frame integration time to millisecond levels. Imaging at three milliseconds per frame the authors could then observe unblurred replisome complexes () and use a similar method of stepwise photobleaching to estimate the stoichiometry of each of the 10 different protein components in the active replisome in a living cell. The most surprising result was that components of the DNA polymerase, a complex molecular motor which runs along the DNA to join new nucleotide bases together as part of the replication process, were present in three copies per replisome (). Earlier experiments under in vitro conditions suggested that only two were present, one sitting on the leading DNA strand the other on the lagging strand. Measurements of the spatial distribution of the components in the replisome suggested that in a minority of cases all three polymerases may be associated with active simultaneous replication, whereas in the majority of cases it is likely that the third polymerase is in a waiting position on the lagging DNA strand.
Conclusions
In the next few years it is likely that functional imaging on single cells at the precision level of single molecules will be utilized as a standard tool in attempts to increase our understanding of the molecular mechanisms underlying biological processes. Other recent studies have already explored many diverse areas of biology including investigations into the dynamics of transcription factors,Citation11 the make-up of functional nanopore complexes responsible for the translocation of proteins across the cell membraneCitation12 and the mobility and architecture of electron-transporting proteins in the oxidative phosphorylation chain.Citation13,Citation14 In addition, fluorescence imaging in vivo now permits measurements to be made on intracellular ion concentrationsCitation15 as well as cell membrane voltagesCitation16 in single functional cells; such experimentation in future combination with single-molecule imaging will present an enormously powerful approach towards addressing fundamental challenges in biological research, as will developments in multi-color single-molecule fluorescence imaging and the use of increasingly complex cellular systems as the experimental model.
Abbreviations
| FLIP | = | fluorescence loss in photobleaching |
| FRAP | = | fluorescence recovery after photobleaching |
| GFP | = | green fluorescent protein |
| mRNA | = | messenger RNA |
| TIRF | = | total-internal-reflection fluorescence |
| YFP | = | yellow fluorescent protein |
Figures and Tables
Figure 1 Fluorescence imaging on single living bacterial cells at the single-molecule level. (A) E. coli cell tethered via a filament in a TIRF field. (B) Sequential brightfield (upper part) and TIRF (lower part) images, cell rotation indicated (yellow cross and arrow), adapted with permission from ref. Citation4. (C) Slimfield microscopy in which a laser beam underfills the back aperture of an objective lens. (D) Brightfield (grey) and slimfield images (yellow) of a single E. coli cell, replisome complexes indicated (arrows). (E) Photobleach intensity trace, raw (blue) and filtered (red) data indicated (adapted with permission from ref. Citation7).
Acknowledgements
M.C.L. is supported by a Royal Society University Research Fellowship, a Hertford College Oxford Research Fellowship, and research grants from the BBSRC (BB/F021224/1) and EPSRC (EP/G061009).
References
- Dobbie IM, Robson A, Delalez, Leake MC. Visualizing single molecular complexes in vivo using advanced fluorescence microscopy. J Vis Exp 2009; 31:1508
- Yu J, Xiao J, Ren X, Lao K, Xie XS. Probing gene expression in live cells, one protein molecule at a time. Science 2006; 311:1600 - 1603
- Jacob F, Monod J. On the regulation of gene activity. Cold Spring Harbor Symp Quant Biol 1961; 26:193 - 211
- Leake MC, Chandler JH, Wadhams GH, Bai F, Berry RM, Armitage JP. Stoichiometry and turnover in single, functioning membrane protein complexes. Nature 2006; 443:355 - 358
- Sowa Y, Berry RM. Bacterial flagellar motor. Q Rev Biophys 2008; 41:103 - 132
- Delalez NJ, Wadhams GH, Rosser G, Xue Q, Brown MT, Dobbie IM, et al. Signal-dependent turnover of the bacterial flagellar switch protein FliM. Proc Natl Acad Sci USA 2010; 107:11347 - 11351
- Reyes-Lamothe R, Sherratt DJ, Leake MC. Stoichiometry and architecture of active DNA replication machinery in Escherichia coli. Science 2010; 328:498 - 501
- Johnson A, O'Donnell M. Cellular DNA replicases: Components and dynamics at the replication fork. Annu Rev Biochem 2005; 74:283 - 315
- McInerney P, Johnson A, Katz F, O'Donnell M. Characterization of a triple DNA polymerase replisome. Mol Cell 2007; 27:527 - 538
- Plank M, Wadhams GH, Leake MC. Millisecond timescale slimfield imaging and automated quantification of single fluorescent protein molecules for use in probing complex biological processes. Integr Biol 2009; 1:602 - 612
- Elf J, Li GW, Xie XS. Probing transcription factor dynamics at the single-molecule level in a living cell. Science 2007; 316:1191 - 1194
- Leake MC, Greene NP, Godun RM, Granjon T, Buchanan G, Chen S, et al. Variable stoichiometry of the TatA component of the twin-arginine protein transport system observed by in vivo single-molecule imaging. Proc Natl Acad Sci USA 2008; 105:15376 - 15381
- Lenn T, Leake MC, Mullineaux CW. Are Escherichia coli OXPHOS complexes concentrated in specialized zones within the plasma membrane?. Biochem Soc Trans 2008; 36:1032 - 1036
- Lenn T, Leake MC, Mullineaux CW. In vivo clustering and dynamics of cytochrome bd complexes in the Escherichia coli plasma membrane. Mol Microbiol 2008; 70:1397 - 1407
- Lo CJ, Leake MC, Berry RM. Fluorescence measurement of intracellular sodium concentration in single Escherichia coli cells. Biophys J 2006; 90:357 - 365
- Lo CJ, Leake MC, Pilizota T, Berry RM. Single-cell measurements of membrane potential, sodium-motive force and flagellar motor speed in Escherichia coli. Biophys J 2007; 93:294 - 302