Abstract
Norbin, a neurite-outgrowth promoting protein, has been found to interact with and regulate several membrane proteins, including metabotropic glutamate receptor 5 (mGluR5). The disruption of both Norbin alleles leads to early embryonic death between 3.5 and 6.5 day post coitus.1 Forebrain specific Norbin knockout (KO) mice are defective in synaptic plasticity,2 an interesting feature considering that Norbin was initially discovered in the context of chemical-induced long term potentiation (LTP),3 a form of synaptic plasticity extensively studied in the context of learning and memory.4 The behavioral phenotypes associated with Norbin conditional KO suggest reduced mGluR5 function. Because of its fundamental functions, Norbin is emerging as a key neuronal regulator. The aim of the present review is to summarize current knowledge about Norbin while emphasizing its role in the nervous system.
Norbin Identification
In a search for novel proteins involved in LTP, rat hippocampal slices treated with a potassium channel blocker (tetraethylammonium) known to induce LTP, were analyzed using a subtractive screening approach and the Norbin gene was found to be upregulated.Citation3 Norbin [neurite-outgrowth related rat brain protein (also called neurochondrin)] is a 75 kDa protein that is evolutionarily conserved from invertebrates (e.g., arthropods, echinoids) to vertebrates (e.g., mammals, amphibians, birds, fishes) () presenting 98% amino acid identity among the rat, mouse and human orthologous proteins. Interestingly, the Norbin primary amino acid sequence does not share any significant homology with other proteins and does not present obvious domains or functional motifs. However, based on secondary structure predictions and sensitive fold recognition tools, it is clear that Norbin is enriched in α-helices (65%) () that are likely organized as tandem repeats.
Norbin Chromosomic Localization and Gene Expression
Norbin gene (Ncdn; MGI:1347351) localizes on mouse chromosome 4D2.1, rat chromosome 5q36 and human chromosome 1p34.3.Citation5,Citation6 It contains 7 exons and codes for two splice variants in mouse and human.Citation6 Several potential binding sites for transcriptional regulators were identified in exon 1 of the Norbin gene.Citation7
Norbin is predominantly expressed in the nervous system.Citation3 Norbin mRNA was first detected embryonically in the hindbrain and spinal cord at day E10.5 by in situ hybridization.Citation8 By day E18, the expression was also detected in forebrain and midbrain. The expression of Norbin is detectable by western blotting at day E18 and increases until day P21, which temporally correspond to the time of dendrite outgrowth.Citation9 In the adult brain, Norbin is highly expressed in the bed nucleus of the stria terminalis (BNST), amygdala and hippocampus and more moderately in septum, piriform cortex, striatum and cortex.Citation2,Citation8 BNST and amygdala are the two components of the brain circuitry regulating fear and anxiety, whereas hippocampus is essential for learning and memory. Norbin is specifically expressed in neurons and presents a somato-dendritic distribution.Citation9 Subcellular fractionation experiments indicated that the majority of Norbin exists in the cytosolic fraction. However, a fraction of Norbin remains insoluble in 1% triton, suggesting a membrane or cytoskeleton association.Citation9 In agreement, Norbin was found to localize to dendritic spines and post-synaptic densities (PSD).Citation2,Citation10
Notably, besides Norbin upregulation in response to LTP induction, Norbin expression was reported to be upregulated in amygdala when a mouse was exposed to the odor of a predator (cat),Citation11 indicating that Norbin's expression is regulated in response to environmental stimuli. In addition, Norbin expression might be differentially regulated in the brain cortex of male and female mice.Citation12
Norbin Interacts with Membrane Proteins and Binds Phospholipids
Norbin was found to interact with several membrane proteins enriched in the brain, including Sema4C,Citation13 several G-protein coupled receptors (GPCRs) from the family A such as the melanin-concentrating hormone receptor 1 (MCHR1)Citation14,Citation15 and from the family C (mGluR1 and mGluR5).Citation2 Truncation and mutagenesis studies indicate that the interactions between Norbin and membrane proteins involve the C-terminal part of Norbin and the membrane-proximal region of the membrane proteins.Citation2,Citation13,Citation14 However, the structural basis of these interactions is not understood. Recently, two sites responsible for Norbin binding were mapped on mGluR5.Citation2 Notably, there is no apparent sequence homology either between those two sites or among other Norbinbinding regions (e.g., Sema4C or MCHR1).
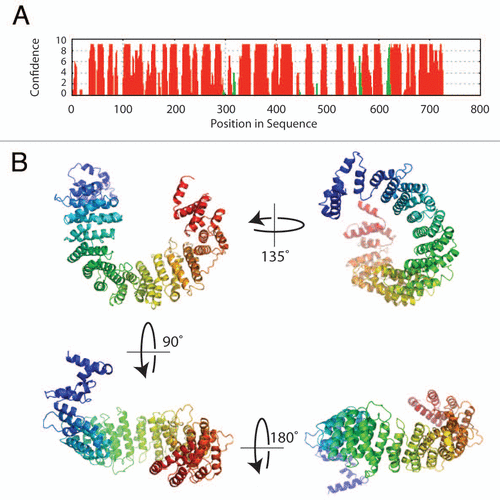
How does Norbin engage different proteins that lack sequence similarity? As mentioned earlier, structural prediction methods suggest that Norbin is almost entirely comprised of α-helical repeat units which show features mostly resemble HEAT repeats (www.embl.de/∼andrade/papers/rep/search.html).Citation16 The crystal structure of HEAT repeats protein importin β1 illustrated that adjacent α-helices stack upon each other to form a right-handed elongated super-helix. The superhelical structure creates a shallow groove that is a protein-protein interaction platform.Citation17 Using comparative modeling methods based on the fold recognition results, we propose a 3D structural model for Norbin (). The concave surface of the curved Norbin helical fold is the likely area dedicated to protein binding. In this eventuality, Norbin would be capable of capturing proteins that present particular structural features instead of relying on primary sequence identity.
Until recently and despite the studies of the physical protein-protein interactions with Norbin, the functional and physiological consequences of interactions involving Norbin remained obscured. It has been reported that Norbin expression increases the steady-state cell surface level of MCHR1 and mGluR5,Citation2,Citation14 suggesting that Norbin plays a key role in the trafficking of transmembrane proteins. Interestingly, mGluR5 C-terminal tail, including the Norbin-binding-region, contains several potential phosphorylation sites.Citation2,Citation18 Phosphorylation of receptor intra-cellular domains is crucial for their endocytosis and recycling.Citation19,Citation20 Norbin, by physically interfering with these modifications, might alter receptor phosphorylation and consequently its trafficking. Along those lines, the central region of Norbin was found to bind to phosphatidic acid, a membrane phospholipid.Citation21 This observation suggests that Norbin may function as an adaptor or scaffold protein linking membrane receptors and lipids, thus facilitating receptor trafficking to the cellular membrane and/or recycling between different cellular membrane compartments.
Involvement of Norbin in Cell Signalingand Neuronal Function
Norbin was recently shown to modulate the signaling pathway of mGluR5.Citation2 mGluR5 primarily responds to the excitatory amino acid neurotransmitter glutamate, localizes at perisynaptic sitesCitation22,Citation23 and plays important roles in normal brain function as well as in several pathological disorders, including schizophrenia.Citation24–Citation26 A recent study indicates that Norbin expression enhances mGluR5 signaling in an in vitro expression system, while the conditional KO of Norbin in transgenic mice reduces mGluR5 function. In line with reduced mGluR5 function, Norbin KO mice show defects in pre-pulse inhibition and psychostimulant induced locomotor activity, two behavioral phenotypes typically observed in rodent models of schizophrenia.Citation2
Norbin has also been shown to modulate the signaling pathway downstream of other Gαq coupled GPCRs such as inhibiting MCHR-induced G-protein activation and downstream calcium influx. It is hypothesized that Norbin, by binding to MCHR1, sterically competes with the binding of G-proteins to the receptor and thereby inhibits G-protein-coupled signal transduction.Citation14 MCHR1 is the melanin-concentrating hormone receptor involved in the regulation of feeding behavior and energy balances.Citation27 Whether Norbin affects metabolic energy through regulating MCHR1 awaits further study.
As mentioned earlier, the expression of Norbin is upregulated during chemical-LTP.Citation3 LTP is the long-lasting enhancement of synaptic strength between neurons, which is an important cellular mechanism underlying learning and memory.Citation4 At least two biochemical phases exist for LTP: an early phase involving modification of existing synaptic proteins and a late phase conveyed by de novo transcription and translation.Citation28 Our recent study showed that forebrain specific Norbin KO abolished both early and late phase hippocampal LTP.Citation2 Consistently, behavioral studies indicated that Norbin deletion impaired spatial learning and memory.Citation29 Activation of mGluR5 is required for hippocampal LTP induction. Considering the positive regulation of mGluR5 by Norbin, it is very likely that Norbin's role in LTP is at least partially dependent upon its interaction with mGluR5. However, the LTP defects seen in Norbin KO mice are slightly more severe than the ones described in mGluR5 KO mice,Citation2,Citation30 suggesting that Norbin might play roles in addition to regulating mGluR5. One obvious possibility is that Norbin deletion also affects mGluR1 function as Norbin also interacts with mGluR1.Citation2
Norbin was found to induce neurite-outgrowth when ectopically overexpressed in cultured neuroblastoma N2a cells. The first 100 amino acids of Norbin contain the activity necessary to induce this phenomenon.Citation3,Citation31 However, the molecular mechanism underlying this neurite-outgrowth promoting activity is not known. Recently, Norbin was found to interact with Dia1, an actin nucleation factor that stimulates barbed end actin filament elongation.Citation31 Although Norbin does not affect the actin polymerizing activity of Dia1, recruiting Dia to a specific sub-cellular localization might contribute to the neurite-outgrowth function of Norbin. Additionally, the capacity of Norbin to direct the trafficking of key membrane receptors toward specific cellular compartments, may also contribute indirectly to neurite outgrowth.
Conclusions
Over the last few years, progress has been made in understanding the function of Norbin in the nervous system. Norbin plays essential roles during development and for synaptic plasticity and interacts physically with several membrane proteins. Structure prediction of Norbin suggests that Norbin may function as a scaffold or adaptor protein that regulates the trafficking and/or signaling of membrane proteins. However, questions are also raised: (1) Is the interaction between Norbin and receptors regulated? Ligand induced GPCR phosphorylation regulates the interaction between GPCRs, GRKs and arrestins.Citation32 As mentioned earlier, Norbin binding domains on mGluR5 include several phosphorylation sites. It will be interesting to test how receptor phosphorylation affects Norbin binding. (2) Could Norbin catalyze receptor crosstalk? Norbin is capable of binding several receptors. Unpublished data indicate that Norbin exists at least partly as a dimer. Assuming that each of the two subunits of a Norbin dimer binds different receptors, Norbin might mediate the coupling and crosstalk among receptors. (3) What is the physiological significance of Norbin interaction with membrane receptors? Norbin interacts with several membrane receptors. With conditional Norbin mice, how Norbin affects receptor function can now be examined by in vivo studies. (4) Is Norbin involved in anxiety/fear responses? Norbin is highly expressed in BNST and amygdala, two brain regions involved in fear and anxiety responses. Forebrain specific Norbin KO mice provide a useful tool to understand this question. In summary, more systematic behavioral studies of Norbin KO mice and future work using molecuar and cellular methods will improve our understanding of Norbin function in the CNS.
Figures and Tables
Figure 1 Phylogenetic analysis of Norbin sequences. Full length Norbin amino acid sequences (FASTA format) were aligned using the program MUSCLE. The phylogenetic analysis was performed on this alignment using PhyML and the tree was drawn using TreeDyn. The GeneID or accession numbers corresponding to each of the sequences used are indicated between parentheses. The mammalian species were grouped (red box) and were analyzed independently to increase the resolution. The class to which the species presented belong is indicated on the right in italics.
Figure 2 Sequence analysis and structure prediction of Norbin. (A) Secondary structure prediction of Norbin. The full length amino acid sequence of Norbin was analyzed using PsiPReD (v2.6). (B) Norbin helical fold architecture. The helical HEAT repeats detected in the Norbin sequence by fold recognition methods were used to build a plausible three-dimensional structure of human Norbin using comparative modeling techniques.Citation16 The seventeen helical repeats shown in (B) (colored in a blue-to-red N-terminal to C-terminal gradient, respectively) form a curved structure with a well defined inner (or concave) surface that forms the likely area of protein interaction.
Acknowledgements
This work was supported in part by grants from the National Institutes of Health (MH40899, DA 10044) (P.G.) and by the award W81XWH-08-1-0111 from the USA Medical Research and Materiel Command to P.G.
References
- Mochizuki R, Dateki M, Yanai K, Ishizuka Y, Amizuka N, Kawashima H, et al. Targeted disruption of the neurochondrin/norbin gene results in embryonic lethality. Biochem Biophys Res Commun 2003; 310:1219 - 1226
- Wang H, Westin L, Nong Y, Birnbaum S, Bendor J, Brismar H, et al. Norbin is an endogenous regulator of metabotropic glutamate receptor 5 signaling. Science 2009; 326:1554 - 1557
- Shinozaki K, Maruyama K, Kume H, Kuzume H, Obata K. A novel brain gene, norbin, induced by treatment of tetraethylammonium in rat hippocampal slice and accompanied with neurite-outgrowth in neuro 2a cells. Biochem Biophys Res Commun 1997; 240:766 - 771
- Bliss TV, Collingridge GL. A synaptic model of memory: long-term potentiation in the hippocampus. Nature 1993; 361:31 - 39
- Mochizuki-Sakisaka R, Dateki M, Ishizuka Y, Yanai K, Matsuda Y, Koga Y, et al. Structural organization of the mouse neurochondrin gene. Int J Mol Med 2004; 14:361 - 366
- Mochizuki R, Ishizuka Y, Yanai K, Koga Y, Fukamizu A, Murakami K. Molecular cloning and expression of human neurochondrin-1 and -2. Biochim Biophys Acta 1999; 1446:397 - 402
- Dateki M, Mochizuki R, Yanai K, Fukamizu A. Identification of the mouse neurochondrin promoter region and the responsible region for cell type specific gene regulation. Neurosci Lett 2004; 356:107 - 110
- Istvánffy R, Vogt Weisenhorn DM, Floss T, Wurst W. Expression of neurochondrin in the developing and adult mouse brain. Dev Genes Evol 2004; 214:206 - 209
- Shinozaki K, Kume H, Kuzume H, Obata K, Maruyama K. Norbin, a neurite-outgrowth-related protein, is a cytosolic protein localized in the somatodendritic region of neurons and distributed prominently in dendritic outgrowth in Purkinje cells. Brain Res Mol Brain Res 1999; 71:364 - 368
- Jordan BA, Fernholz BD, Boussac M, Xu C, Grigorean G, Ziff EB, et al. Identification and verification of novel rodent postsynaptic density proteins. Mol Cell Proteomics 2004; 3:857 - 871
- Koks S, Luuk H, Nelovkov A, Areda T, Vasar E. A screen for genes induced in the amygdaloid area during cat odor exposure. Genes Brain Behav 2004; 3:80 - 89
- Mani ST, Kumar RC, Thakur MK. Age- and sex-related expression of norbin in the brain cortex of mice. Neurosci Lett 2001; 308:57 - 59
- Ohoka Y, Hirotani M, Sugimoto H, Fujioka S, Furuyama T, Inagaki S. Semaphorin 4C, a transmembrane semaphorin, associates with a neurite-outgrowth-related protein, SFAP75. Biochem Biophys Res Commun 2001; 280:237 - 243
- Francke F, Ward RJ, Jenkins L, Kellett E, Richter D, Milligan G, et al. Interaction of neurochondrin with the melanin-concentrating hormone receptor 1 interferes with G protein-coupled signal transduction but not agonist-mediated internalization. J Biol Chem 2006; 281:32496 - 32507
- Ward RJ, Jenkins L, Milligan G. Selectivity and functional consequences of interactions of family A G protein-coupled receptors with neurochondrin and periplakin. J Neurochem 2009; 109:182 - 192
- Zhang Y. Progress and challenges in protein structure prediction. Curr Opin Struct Biol 2008; 18:342 - 348
- Andrade MA, Petosa C, O'Donoghue SI, Müller CW, Bork P. Comparison of ARM and HEAT protein repeats. J Mol Biol 2001; 309:1 - 18
- Lee JH, Lee J, Choi KY, Hepp R, Lee JY, Lim MK, et al. Calmodulin dynamically regulates the trafficking of the metabotropic glutamate receptor mGluR5. Proc Natl Acad Sci USA 2008; 105:12575 - 12580
- Marchese A, Paing MM, Temple BR, Trejo J. G protein-coupled receptor sorting to endosomes and lysosomes. Annu Rev Pharmacol Toxicol 2008; 48:601 - 629
- Tobin AB, Butcher AJ, Kong KC. Location, location, location…site-specific GPCR phosphorylation offers a mechanism for cell-type-specific signalling. Trends Pharmacol Sci 2008; 29:413 - 420
- Ktistakis NT, Delon C, Manifava M, Wood E, Ganley I, Sugars JM. Phospholipase D1 and potential targets of its hydrolysis product, phosphatidic acid. Biochem Soc Trans 2003; 31:94 - 97
- Abe T, Sugihara H, Nawa H, Shigemoto R, Mizuno N, Nakanishi S. Molecular characterization of a novel metabotropic glutamate receptor mGluR5 coupled to inositol phosphate/Ca2+ signal transduction. J Biol Chem 1992; 267:13361 - 13368
- Romano C, Sesma MA, McDonald CT, O'Malley K, Van den Pol AN, Olney JW. Distribution of metabotropic glutamate receptor mGluR5 immunoreactivity in rat brain. J Comp Neurol 1995; 355:455 - 469
- Nakanishi S, Nakajima Y, Masu M, Ueda Y, Nakahara K, Watanabe D, et al. Glutamate receptors: brain function and signal transduction. Brain Res Brain Res Rev 1998; 26:230 - 235
- Bear MF, Huber KM, Warren ST. The mGluR theory of fragile X mental retardation. Trends Neurosci 2004; 27:370 - 377
- Conn PJ, Christopoulos A, Lindsley CW. Allosteric modulators of GPCRs: a novel approach for the treatment of CNS disorders. Nat Rev Drug Discov 2009; 8:41 - 54
- Lembo PM, Grazzini E, Cao J, Hubatsch DA, Pelletier M, Hoffert C, et al. The receptor for the orexigenic peptide melanin-concentrating hormone is a G-protein-coupled receptor. Nat Cell Biol 1999; 1:267 - 271
- Kandel ER. The molecular biology of memory storage: a dialog between genes and synapses. Biosci Rep 2001; 21:565 - 611
- Dateki M, Horii T, Kasuya Y, Mochizuki R, Nagao Y, Ishida J, et al. Neurochondrin negatively regulates CaMKII phosphorylation and nervous system-specific gene disruption results in epileptic seizure. J Biol Chem 2005; 280:20503 - 20508
- Lu YM, Jia Z, Janus C, Henderson JT, Gerlai R, Wojtowicz JM, et al. Mice lacking metabotropic glutamate receptor 5 show impaired learning and reduced CA1 long-term potentiation (LTP) but normal CA3 LTP. J Neurosci 1997; 17:5196 - 5205
- Schwaibold EM, Brandt DT. Identification of Neurochondrin as a new interaction partner of the FH3 domain of the Diaphanous-related formin Dia1. Biochem Biophys Res Commun 2008; 373:366 - 372
- Gainetdinov RR, Premont RT, Bohn LM, Lefkowitz RJ, Caron MG. Desensitization of G protein-coupled receptors and neuronal functions. Annu Rev Neurosci 2004; 27:107 - 144