Abstract
We have recently reported the isolation and characterization of Plasmodium falciparum Dbp5/DDX19 homologue PfD66 and the results indicate that it contains ATP-dependent bipolar DNA and RNA unwinding activity, intrinsic nucleic acid-dependent ATPase and RNA-binding activities (Molecular and Biochemical Parasitology, (In press) 10.1016/j.molbiopara.2010.12.003). In the present study we report the effect of a number of compounds such as actinomycin D, aphidicolin, camptothecin, cyclophosphamide, 4’,6’-di-amidino-2-phenylindole (DAPI), daunorubicin, distamycin, ethidium bromide, ellipticine, genistein, mitoxantrone, nalidixic acid, netropsin, nogalamycin, novobiocin and VP-16 on the DNA unwinding and ATPase activities of PfD66. The results indicate that DAPI, ethidium bromide, netropsin and nogalamycin efficiently inhibited the helicase and ATPase activities of PfD66. These studies will make an important contribution in understanding the mechanism of DNA unwinding by Plasmodium falciparum helicase PfD66.
Malaria is a devastating disease and a scourge to mankind since antiquity, which affects 300–500 million people worldwide and results in an estimated 880,000 deaths annually.Citation1,Citation2 Malaria is a disease of global concern causing major challenges to health and socio-economic development in more than 100 countries of the world. Plasmodium falciparum, Plasmodium ovale, Plasmodium malariae and Plasmodium vivax are the main species that cause malaria in human.Citation1,Citation2 Among the species of Plasmodium commonly known to infect humans, Plasmodium falciparum is the most virulent and causes the most fatal form of malarial infection resulting in 95% of all malaria-related deaths. In the absence of clinically proven malaria vaccine, there are only a limited number of affordable drugs available for the purpose of treatment.Citation2,Citation3 Moreover, the resurgence in the resistance of the parasite to both chloroquine and sulphadoxine pyremethamine in most malaria affected areas along with the spread of the disease into new geographic regions has led to doubling in the number of deaths in many parts of the world.Citation1,Citation4–Citation6 The search for affordable and novel pharmacophores for the purpose of malaria intervention requires identification of new chemotherapeutic targets. The completed genome of Plasmodium falciparum has opened new avenues for research as it will help to identify key targets in biochemical pathways, which are parasite-specific and can be used as potential drug targets.Citation7,Citation8 Many of the proteins of the parasite may be good targets for the development of antimalarial drugs. The universal presence of helicases in eukaryotes, prokaryotes, viruses and bacteriophages reflects their fundamental importance in all aspects of DNA and RNA metabolism. Recently we have reported the genome-wide analysis of helicases from Plasmodium falciparum.Citation9 The DEAD/H-box containing RNA helicases are a family of enzymes that can modify and restructure RNA and RNPs in ATP-dependent manner.Citation9 Since helicases play essential roles in the metabolism of DNA and RNA, helicase inhibitors may offer a feasible route towards the development of novel drugs.Citation10 Recently we have reported the detailed characterization of the DDX19/Dbp5 homolog designated as PfD66 from Plasmodium falciparum.Citation11 In the present study we have characterized PfD66 further by using certain compounds that block the helicase and DNA-dependent ATPase activities of this enzyme. The inhibition of enzyme activities could be due to the formation of a compound-DNA complex that hampers the movement of the enzyme.
Antihelicase activity of a variety of DNA intercalating agents was checked using the standard strand displacement helicase assay.Citation11 Three classes of nucleic acid interacting agents used in the study were: (1) DNA-intercalating agents, such as daunorubicin, ethidium bromide, ellipticine, nogalamycin, cyclophosphamide, mitoxantrone and actinomycin D, (2) minor groove binders, such as distamycin and netropsin and (3) non-intercalating topoisomerase inhibitors, such as camptothecin, VP-16, novobiocin and DNA polymerase alpha inhibitor like aphidicolin. The effect of these ligands on the DNA unwinding activity of PfD66 (20 nM) was examined by including 50 µM of each ligand separately in the standard helicase assay using 1 ng of the hanging tails substrate (). The results indicated that compounds like nalidixic acid, ethidum bromide, DAPI, netropsin, daunorubicin and noglamycin inhibited the activity of PfD66 effectively (, lanes 6, 7, 9, 10, 14 and 15 respectively) at 50 µM concentration. Whereas the other compounds like aphidicolin, actinomycin D, camptothecin, cyclophosphamide, mitoxantrone, novobiocin, VP-16, ellipticine and genistein (, lanes 2–5, 8, 11–13 and 16 respectively) were unable to inhibit the DNA-unwinding activity of PfD66 at 50 µM concentration.
The compounds, which inhibited the helicase activity, were investigated further for the kinetics of inhibition. For this, each inhibitor was included in the helicase reaction at the final concentration ranging from 0.25 to 20 µM. The results of these inhibitors are shown in . The apparent IC50 was calculated for each of the inhibitors from the data. The most effective inhibitors were netropsin and ethidium bromide, with apparent IC50 values of 0.5 µM and 1 µM respectively () followed by DAPI and nogalamycin with apparent IC50 values of 1.9 and 5 µM respectively ().
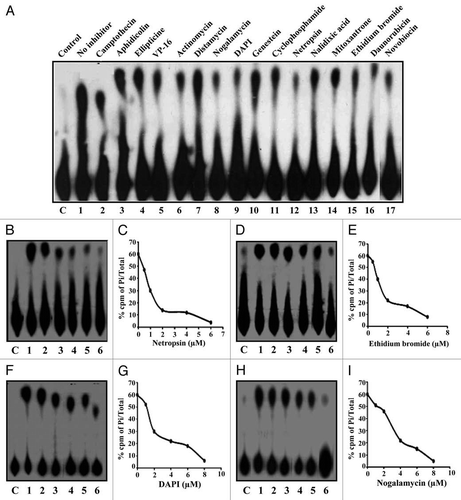
For studying the effect of DNA intercalating compounds described above on the ssDNA dependent ATPase activity, different compounds were added to the reaction mixture prior to the addition of the enzyme. The effect of the compounds on the ssDNA dependent ATPase activity of PfD66 was studied by including 50 µM of the compound in each case in a standard ATPase assay as described previously in reference Citation11. The results showed that nogalamycin, DAPI, netropsin and ethidium bromide (, lanes 8, 9, 12 and 15 respectively) showed significant inhibition of the ssDNA dependent ATPase activity of PfD66. The compounds like camptothecin, aphidicolin, ellipticine, VP-16, actinomycin D, distamycin, genestein, cyclophosphamide, nalidixic acid, mitoxantrone, daunorubicin and novobiocin (, lanes 2–7, 10, 11, 13, 14, 16 and 17 respectively) were not able to inhibit the ssDNA-dependent ATPase activity of PfD66 significantly at 50 µM concentration.
The compounds, which inhibited the ssDNA-dependent ATPase activity, were investigated further for the kinetics of inhibiton. Each inhibitor was included in the reaction at the final concentration ranging from 0.5 to 10 µM and the results of the kinetics of these inhibitors are shown in . The results show that the most effective inhibitor is netropsin with an apparent IC50 value of 1 µM ( and C), followed by ethidium bromide, DAPI and nogalamycin with the IC50 values of 1.5 µM, 2 µM and 3.2 µM respectively (). It was concluded from the inhibitor studies that netropsin was the most potent inhibitor of PfD66 having the lowest Ki values for helicase and ATPase activities.
RNA helicases represent a large family of proteins implicated in many biological processes including ribosome biogenesis, splicing, translation and mRNA degradation. However, these proteins have little substrate specificity, making inhibition of selected helicases a challenging problem.Citation12 Recently a coral derived natural product, hippuristanol was found to be an inhibitor of eIF4A.Citation12 Whereas, typically it has been found that known nucleic acid binding agents disrupt helicase activity by directly binding to the nucleic acid.Citation13 Moreover, the DEAD-box helicases are known to bind nucleic acids more efficiently in the presence of ATP and vice versa.Citation14–Citation16 Therefore any significant reduction in nucleic acid binding and further unwinding might lead to a concomitant decrease in the ATPase activities of the enzyme as well. Therefore, it will be interesting to note the effect of these intercalating agents on the helicase activity of RNA helicases. The effect of these nucleic acid binding agents on inhibition of unwinding of the duplex might have a deleterious effect on further DNA/RNA transaction processes.Citation10,Citation17
The effective DNA-binding ligands, which inhibited the DNA unwinding and ATPase reactions catalyzed by PfD66 were DAPI, nogalamycin, ethidium bromide and netropsin. An oligopyrrolamidine, netropsin binds to the minor groove of DNA and inhibits WRN and BLM helicases but it did not inhibit E. coli UvrD helicase activity.Citation18 Nogalamycin and daunorubicin also interact with DNA and the inhibition by noglamycin depends on the source of the enzyme and is highly variable because the IC50 value for this compound ranged from 0.1 to >650 µM for various viral helicases.Citation19 These anthracyclines, nogalamycin and daunorubicin are well known universal inhibitors of most helicases such as HDHII, SV40 large T antigen, PcDDH45, PfDH60 and the viral helicase of the Flaviridae family.Citation17,Citation19–Citation21 The mechanism by which these ligands inhibit unwinding reaction is through intercalation into the duplex DNA substrate. This probably provides a physical block to continued translocation of the helicase, resulting into the inhibition. The inhibition of ATPase activity may be explained in terms of formation of ternary complex (enzyme/DNA/inhibitor). Among all the inhibitors studied, netropsin was the most potent inhibitor having the minimum Ki value for helicase and the ATPase activities. The findings reported in the present study should make a valuable contribution in unravelling the mechanism by which the DNA-interacting compounds act to inhibit the activities of the malaria parasite helicase.
Figures and Tables
Figure 1 The effect of different DNA intercalating compounds on the DNA unwinding activity of PfD66. The reaction was performed using purified enzyme PfD66 and 50 µM of the compound. The DNA-interacting compounds used in the present study were purchased from Topogene Inc., (Columbus, OH), Sigma Chemical Co., (St. Louis, MO) and BDH (E. Merck, Mumbai, India). All of these compounds were dissolved in dimethylsulfoxide (DMSO). The compound added is mentioned at the top of the autoradiogram and (C) is control, no inhibitor is control having enzyme without inhibitor and (B) is the heat denatured substrate. (B–I) Concentration curves of netropsin (B), ethidium bromide (D), DAPI (F) and nogalamycin (H). The DNA helicase reactions were performed by using the same substrate and enzyme as in (A) in the presence of increasing concentration of various compounds. In each (C) is control without enzyme and (B) is the boiled substrate and lanes 1–6 are reactions in the presence of increasing amounts of various compounds. (C, E, G and I) show the quantitative data.
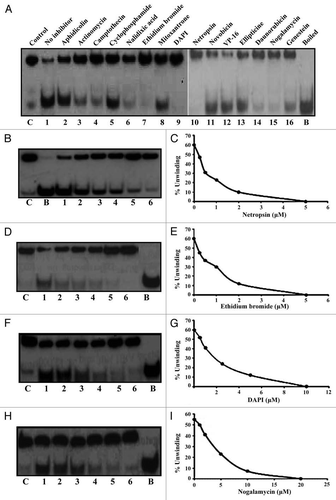
Figure 2 The effect of different DNA intercalating compounds on the ATPase activity of PfD66. The reaction was performed using purified enzyme and 50 µM of the compound. The compound added is mentioned at the top of the autoradiogram and (C) is no enzyme control and lane 1 is control having enzyme without inhibitor. (B–I) Concentration curves of netropsin (B), ethidium bromide (D), DAPI (F) and nogalamycin (H). The assays were performed by using the same substrate and enzyme as in (A) in the presence of increasing concentration of various compounds. In each (C) is control without enzyme and lanes 1–6 are reactions in the presence of increasing amounts of various compounds. (C, E, G and I) show the quantitative data.
Acknowledgements
J.M. is supported by fellowship from the Department of Biotechnology, Govt. of India. This work in R.T.'s laboratory is partially supported by the Department of Biotechnology and Defence Research and Development Organization grants. Infrastructural support from the Department of Biotechnology, Government of India is gratefully acknowledged.
Addendum to:
References
- Snow RW, Guerra CA, Noor AM, Myint HY, Hay SI. The global distribution of clinical episodes of Plasmodium falciparum malaria. Nature 2005; 434:214 - 217
- Tuteja R. Malaria-An overview. FEBS J 2007; 274:4670 - 4679
- Todryk SM, Hill AV. Malaria vaccines: the stage we are at. Nat Rev Microbiol 2007; 5:487 - 489
- White NJ. Preventing antimalarial drug resistance through combinations. Drug Resist Updat 1998; 1:3 - 9
- Trape JF. The public health impact of chloroquine resistance in Africa. Am J Trop Med Hyg 2001; 64:12 - 17
- Sachs J, Malaney P. The economic and social burden of malaria. Nature 2002; 415:680 - 685
- Carlton JM, Angiuoli SV, Suh BB, Kooij TW, Pertea M, Silva JC, et al. Genome sequence and comparative analysis of the model rodent malaria parasite Plasmodium yoelii yoelii. Nature 2002; 419:512 - 519
- Gardner MJ, Shallom SJ, Carlton JM, Salzberg SL, Nene V, Shoaibi A, et al. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature 2002; 419:498 - 511
- Tuteja R. Genome wide identification of Plasmodium falciparum helicases: a comparison with human host. Cell Cycle 2010; 9:104 - 120
- Tuteja R. Helicases: feasible anti-malarial drug target for Plasmodium falciparum. FEBS J 2007; 274:4699 - 4704
- Mehta J, Tuteja R. A novel dual Dbp5/DDX19 homologue from Plasmodium falciparum requires Q motif for activity. Mol Biochem Parasitol 2011; 176:58 - 63
- Lindqvist L, Oberer M, Reibarkh M, Cencic R, Bordeleau ME, Vogt E, et al. Selective pharmacological targeting of a DEAD box RNA helicase. PLoS One 2008; 13:1583
- George JW, Ghate S, Matson SW, Besterman JM. Inhibition of DNA helicase II unwinding and ATPase activities by DNA-interacting ligands. Kinetics and specificity. J Biol Chem 1992; 267:10683 - 10689
- Lorsch JR, Herschlag D. Kinetic dissection of fundamental processes of eukaryotic translation initiation in vitro. EMBO J 1998; 18:6705 - 6717
- Polach KJ, Uhlenbeck OC. Cooperative binding of ATP and RNA substrates to the DEAD/H protein DbpA. Biochemistry 2002; 41:3693 - 3702
- Cordin O, Tanner NK, Doere M, Linder P, Banroques J. The newly discovered Q motif of DEAD-box RNA helicases regulates RNA-binding and helicase activity. EMBO J 2004; 23:2478 - 2487
- Pradhan A, Tuteja R. Plasmodium falciparum DNA helicase 60: dsRNA- and antibody-mediated inhibition of the malaria parasite growth and downregulation of its enzyme activities by DNA-interacting compounds. FEBS J 2006; 273:3545 - 3556
- Brosh RM Jr, Karow JK, White EJ, Shaw ND, Hickson ID, Bohr VA. Potent inhibition of werner and bloom helicases by DNA minor groove binding drugs. Nucleic Acids Res 2000; 28:2420 - 2430
- Borowski P, Niebuhr A, Schmitz H, Hosmane RS, Bretner M, Siwecka MA, et al. NTPase/helicase of Flaviviridae: inhibitors and inhibition of the enzyme. Acta Biochim Pol 2002; 49:597 - 614
- Tuteja N, Phan TN, Tuteja R, Ochem A, Falaschi A. Inhibition of DNA unwinding and ATPase activities of human DNA helicase II by chemotherapeutic agents. Biochem Biophys Res Commun 1997; 236:636 - 640
- Tuteja R, Tuteja N, Malhotra P, Chauhan VS. Replication fork stimulated eIF-4A from Plasmodium cynomolgi unwinds DNA in the 3′ to 5′ direction and is inhibited by DNA-interacting compounds. Arch Biochem Biophys 2003; 414:108 - 114