Abstract
The centrosome is a complex cell organelle in higher eukaryotic cells that functions in microtubule organization and is integrated into major cellular signaling pathways.1-3 For example a tight link exists between cell cycle regulation and centrosome duplication, as centrosome number must be precisely controlled to ensure high fidelity of chromosome segregation.4 The analysis of the centrosome's protein composition provides the opportunity for a better understanding of centrosome function and to identify possible links to cellular signaling pathways.5,6 Our proteomics study of the Drosophila centrosome recently identified 251 centrosome candidate proteins that we subsequently characterized by RNAi in Drosophila SL2 cells and classified according to their function in centrosome duplication/segregation, structure maintenance and cell cycle regulation.7 Interestingly, functional characterization of their human orthologue proteins revealed the highest functional conservation in the process of centrosome duplication and separation. To analyze functional and biochemical interdependencies further, we carried out an analysis of the gene ontology (GO) annotation of the identified Drosophila centrosome proteins, as well as of the human centrosome proteome.5 The GO analysis of the group of proteins that did not show a centrosome, chromosome segregation or cell cycle related phenotype in our RNAi assays suggests that these molecules may constitute linker proteins to other cellular signaling pathways. Furthermore, the results of our GO analysis of components of the human and of the Drosophila centrosome reflect the somatic and embryonic origin, respectively, of the isolated centrosomes, implicating the Drosophila centrosome proteins in developmental signaling and cell differentiation.
GO Analysis of Different Functional Classes of Drosophila Centrosome Proteins Reveals Distinct Ontology Groups
To reveal similarities or differences between distinct functional groups in the centrosomal proteome, we analyzed the enrichment of GO terms using the DAVID Functional Annotation Tool (david.abcc.ncifcrf.gov/).Citation8,Citation9 Enrichment of biological process level 3 annotations was tested for groups of proteins affecting (1) centrosome duplication and separation, (2) centrosome structure and (3) proteins that showed no phenotype related to centrosome, cell cycle or chromosome segregation in our functional analysis of MS identified centrosome candidate proteins (). The enriched GO annotations of the proteins implicated in centrosome duplication/separation and structure maintenance partly overlap, with members of both groups being implicated in cell cycle processes and microtubule cytoskeleton and spindle organization, respectively. GO terms that relate to meiosis or microtubule organizing center organization were exclusively enriched in the centrosome structure maintenance group, whereas proteins functioning in centrosome duplication and separation shared the terms protein biosynthesis, metabolism and complex assembly as well as microtubule-based movement, as expected.
The centrosomal candidate proteins that showed no phenotype in the functional analysis are not overlapping with either of the two other phenotypic groups. The enrichment of terms such as tissue development or transport in the “no phenotype” group reflects an involvement of those proteins in biological processes not directly related to those specific for centrosome duplication/separation or structure proteins. This might indicate that this subset of centrosomal proteins forms a distinct group that links the centrosome to other pathways or processes without having a function in centrosome key events like duplication, separation or structure maintenance.
Semantic Similarity of Protein Sets
The functional similarity of a set of gene products can be measured based on the Gene Ontology (GO) annotations associated with them. We used the method of Wang et al.Citation10 to quantify similarity scores within and between sets of functionally related proteins. For a given protein pair, the algorithm computes a similarity score between 0 and 1, taking into account the network topology of the GO database. Thus, for functionally unrelated proteins the score tends towards 0 and for related proteins the score tends towards 1.
For each of the three protein groups defined by their RNAi-mediated phenotype (structure, duplication/segregation, no phenotype), we calculated the average semantic similarity score (). The group of proteins without centrosome phenotype upon RNAi was found to have an overall score of 0.30, which indicates a rather low functional similarity of its participants. Yet for the other two protein groups associated with duplication/segregation and structure related phenotypes, the score was clearly higher (0.51 and 0.48, respectively). This emphasizes the assumption that each of these two groups consists of proteins that share similar functions.
We also calculated the semantic similarity between the three groups (). When comparing the structure or duplication/segregation group with the “no phenotype” group, moderate scores of 0.34 were found. In contrast, comparison between the structure and the duplication/segregation group resulted in a higher score (0.49). This suggests an interrelation of the two protein groups, which is in line with the hypothesis that centrosome structure maintenance and centrosome duplication are associated cellular processes.
GO Term Enrichment Analysis of Human vs. Drosophila Centrosome Proteome
Comparison between the DrosophilaCitation7 and the human centrosome proteomeCitation5 revealed an overlap of only ∼45% and led to the question whether the differences between the two datasets are due to the source/method of preparation of the centrosomes or whether indeed non-homologous proteins fulfill similar centrosome-related functions in the two species. If the latter hypothesis was true, then non-homologous proteins would be involved in the same centrosome-regulatory processes in the two species. To test this hypothesis, we performed a comparative analysis of GO term enrichment between the proteomic datasets (). At first glance it is surprising that only ∼17% of the GO terms were enriched in both datasets. These terms, however, are rather general and are associated with ∼73% of all human and ∼58% of all Drosophila proteins analyzed. Interestingly, centriolar proteins or those directly associated with centrioles, such as Sas6, Centrin2, Centrobin or PLK4,Citation11–Citation13 make up a large portion (56%) of the factors associated with terms specific for human centrosome proteins. This finding is consistent with the fact that the human centrosomes were isolated from cells primarily in interphase of the cell cycle, in which the relative centrosomal abundance of centrioles is significantly higher than in mitotic centrosomes rich in pericentriolar material (PCM). The remaining 44% of this group are primarily microtubule associated-(EB1, CLASP1, LIS1) and PCM proteins (NDE1, CEP120, 14-3-3 ε). In conclusion, approximately 2/3 of each of the two different centrosome proteomes are involved in major processes of microtubule cytoskeleton organization and cell cycle regulation and about 50% of these have no known orthologs in the respective other dataset. This suggests that a significant portion of these important functions is mediated by different proteins in the two species. The annotations of roughly 1/3 of the proteomes do not overlap and this is likely due to the different sources of the centrosomes, as discussed above.
Figures and Tables
Figure 1 (A) The enrichment of GO annotation terms (biological process level 3) in the phenotypic classes “centrosome duplication/separation.” “Centrosome structure” and “no phenotype” were analyzed using DAVID Functional Annotation Tool with the 251 MS-identified centrosomal candidate proteins as background. Bars indicate the fold enrichment of the respective GO terms in the phenotypic classes. (B) Semantic similarity scores (i) within and (ii) between the three protein sets. Scores were calculated with the Bioconductor package ‘GOSemSim’ (Version 1.8.2). N gives the total number of proteins, N effective gives the number of proteins with GO annotation sufficient for score calculation. p values for score differences were calculated using the Mann-Whitney U test.
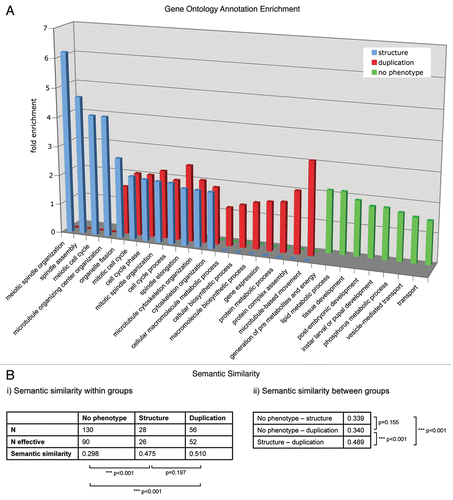
Figure 2 Comparative GO term enrichment analysis reveals functional homology between the Drosophila and human centrosome proteome. GO term enrichment in the category biological process level 3 was analyzed for all proteins classified as human centrosomal, centrosome candidate and centrosome novelCitation5 using the DAVID bioinformatics tool and the whole human genome as the background. The Drosophila centrosome proteome was analyzed accordingly and statistically significantly enriched terms (p value < 0.01) were compared between the two datasets. Less informative terms not directly related to centrosome structure or function were merged and labeled with the term of the next higher level of the GO hierarchy (italic).

Acknowledgements
This work was supported by (a) B.L. laboratory: Berliner Senat für Kultur, Wissenschaft und Forschung, EFRE; NGFN Plus, IG Mutanom; EU. (b) A.P. laboratory: Leibniz Society, Pakt für Forschung und Innovation. (c) R.H. laboratory: German Science Foundation grant No. HE4607/3-1.
Addendum to:
References
- Lange BMH. Integration of the centrosome in cell cycle control, stress response and signal transduction pathways. Curr Opin Cell Biol 2002; 14:35 - 43
- Doxsey S. Re-evaluating centrosome function. Nat Rev Mol Cell Biol 2001; 2:688 - 698
- Nigg EA, Raff JW. Centrioles, centrosomes and cilia in health and disease. Cell 2009; 139:663 - 678
- Ganem NJ, Godinho SA, Pellman D. A mechanism linking extra centrosomes to chromosomal instability. Nature 2009; 460:278 - 282
- Andersen JS, Wilkinson CJ, Mayor T, Mortensen P, Nigg EA, Mann M. Proteomic characterization of the human centrosome by protein correlation profiling. Nature 2003; 426:570 - 574
- Bettencourt-Dias M, Glover DM. Centrosome biogenesis and function: centrosomics brings new understanding. Nat Rev Mol Cell Biol 2007; 8:451 - 463
- Müller H, Schmidt D, Steinbrink S, Mirgorodskaya E, Lehmann V, Habermann K, et al. Proteomic and functional analysis of the Drosophila centrosome. EMBO J 2010; 29:3344 - 3357
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009; 4:44 - 57
- Dennis G Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, et al. DAVID: Database for Annotation, Visualization and Integrated Discovery. Genome Biol 2003; 4:3
- Wang JZ, Du Z, Payattakool R, Yu PS, Chen CF. A new method to measure the semantic similarity of GO terms. Bioinformatics 2007; 23:1274 - 1281
- Habedanck R, Stierhof YD, Wilkinson CJ, Nigg EA. The Polo kinase Plk4 functions in centriole duplication. Nat Cell Biol 2005; 7:1140 - 1146
- Zou C, Li J, Bai Y, Gunning WT, Wazer DE, Band V, et al. Centrobin: a novel daughter centriole-associated protein that is required for centriole duplication. J Cell Biol 2005; 171:437 - 445
- Salisbury JL. Centrin, centrosomes and mitotic spindle poles. Curr Opin Cell Biol 1995; 7:39 - 45