Abstract
Biochemical assessment of channel structure/function is incredibly challenging. Developing computational tools that provide these data would enable translational research, accelerating mechanistic experimentation for the bench scientist studying ion channels. Starting with the premise that protein sequence encodes information about structure, function and evolution (SF&E), we developed a unified framework for inferring SF&E from sequence information using a knowledge-based approach. The Gestalt Domain Detection Algorithm-Basic Local Alignment Tool (GDDA-BLAST) provides phylogenetic profiles that can model, ab initio, SF&E relationships of biological sequences at the whole protein, single domain, and single-amino acid level.1,2 In our recent paper,4 we have applied GDDA-BLAST analysis to study canonical TRP (TRPC) channels1 and empirically validated predicted lipid-binding and trafficking activities contained within the TRPC3 TRP_2 domain of unknown function. Overall, our in silico, in vitro, and in vivo experiments support a model in which TRPC3 has signal-sensing antennae which are adorned with lipid-binding, trafficking, and calmodulin regulatory domains. In this Addendum, we correlate our functional domain analysis with the cryo-EM structure of TRPC3.3 In addition, we synthesize recent studies with our new findings to provide a refined model on the mechanism(s) of TRPC3 activation/deactivation.
Correlating Computational Models to Experimental Evidence
In our recent theoretical paper,Citation1 we have proposed that phylogenetic profiles built using ontologically related functional domain profiles provide a unified framework for deriving SF&E information from protein sequences. We applied this approach to study transient receptor potential channels (TRP) as a benchmark set of proteins.Citation4 Overall, functional annotation for TRP channels is lacking. For example, formal queries of TRPC protein sequences with popular domain detection algorithms (Pfam, SMART, NCBI-CDD, Interproscan) predict that TRPC channels contain only one functional domain (ion channel), and two domains whose function is unknown (ankyrin, TRP_2), thus leaving the majority of the protein without functional annotation. This is in stark contrast to the literature in which multiple protein and lipid interactions,Citation5–Citation12 as well as sites for covalent modification have been identified.Citation13–Citation17
In , we present our computational analysis of human TRPC3 with multiple structural/functional profiles related by ontology (i.e., keywords) for peripheral lipid-binding, integral lipid-binding, calmodulin-binding and ankyrin repeats. The results identify multiple lipid/calmodulin-binding modules along the cytosolic N and C termini. Indeed, 4/5 regions have been experimentally validated to bind lipids in TRP channels.Citation9,Citation11,Citation18,Citation19 In our recent manuscript,Citation4 we validated GDDA-BLAST predictions in the TRP_2 domain of TRPC3 for lipid-binding/trafficking activities. Indeed, TRP_2 has relatively non-specific lipid-binding activity, which upon mutation of S209/S213 to mirror TRPC1, increases its lipid-binding affinity. This is associated with defective channels unable to insert into the plasma membrane in response to diacylglycerol (DAG)-mediated membrane destabilization. Furthermore, based on (i) our analysis of trafficking motifs in TRPC3 (which are comprised primarily of SNARE domain profiles),Citation1 and (ii) the role of TRPC3 TRP_2 in its own vesicle fusion,Citation4 we suggest that TRPC3 contains “SNARE-like” activity, similar to that observed for TRPM7.Citation20 Taken together, our recent findings provide a mechanistic and functional difference between DAG-sensitive TRPCs (3/6/7) and DAG-insensitive TRPCs (1/4/5), which has been a controversial question in the TRPC field for almost a decade.Citation21,Citation22
A recent study by Mio et al. obtained a cryo-EM structure of Transient Receptor Potential Channel 3 (TRPC3) and modeled the six transmembrane helices with the atomic structure of the potassium channels KcsA and Kv1.2,Citation3 (). Interestingly, these authors also determined that TRPC3 contains a globular, and presumably hydrophobic, inner-shell surrounded by signal sensing antenna derived from the cytosolic N and C-termini. Indeed, GDDA-BLAST predicts the inner-shell in TRPC3/6/7 as well as signal sensing antennae with the capacity to make multiple lipid contacts with the plasma membrane (i.e., swelling). Based on our computational functional domain predictions we have provided a rough annotation on the cryo-EM structure. Although speculative, it is intriguing how GDDA-BLAST results correlate to known structural elements.Citation3,Citation23 A number of implications can be derived from such a model. For example, the N-terminus forms a “thumb” on the ankyrin repeat “hand”. Further, the C-terminus forms an “arm” perpendicular to the plasma membrane that could be parallel when bound to PIP2 (TRP-box) and/or PIP3 (C-terminal arm).Citation11,Citation19 Two regions between the ankyrin repeats and the inner-shell are not obvious as to their relative amino acid positions. Nevertheless, we propose that the linker (green) comprises the TRP_2 domain, and that the “knob-like projection” (turquoise) near the plasma membrane is a lipid-binding domain.
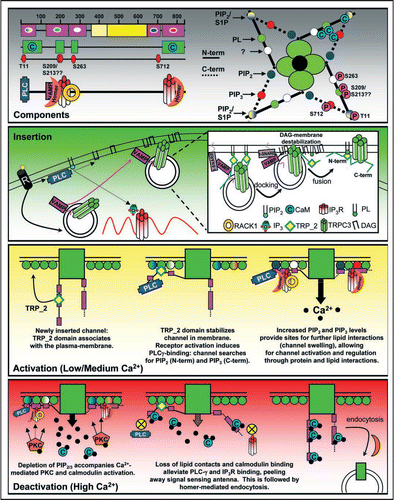
In , we present here a model for the activation/deactivation mechanism of TRPC3, which combines our computational predictions and biochemical/cell biological reports from the literature. Fundamental to this model is the hypothesis that the N and C-termini of TRPC3 are signal-sensing antennae, which can engage and disengage with the membrane. Further, we propose a scheme by which these antennae can be regulated (in a graded manner) by lipids, calmodulin and PKC/PKG phosphorylation. Important points in this model include the VAMP and TRP_2 dependent forward trafficking/insertion of TRPC3, which is facilitated by DAG-mediated membrane destabilization. Although DAG has been implicated as a “direct activator” for TRPC3/6/7,Citation21,Citation22 the experimental data from these and other manuscripts are congruent with our results, as none of these reports measured the plasma membrane expression of these channels in their experimentation.
Once inserted, we envision a step-wise recruitment/location of the channel to regions, which are rich in PIP2 and PIP3. Finding micro-domains containing these lipids may be facilitated by protein-protein interactions such as phospholipase Cγ(PLCγ),Citation19 receptor for activated c kinase (RACK1),Citation6 inositol trisphosphate receptor (IP3R)Citation24 and/or homer.Citation25,Citation26 Once properly localized we propose that the lipid-interactions, in part, control channel activity as has been previously demonstrated. We suggest that, in a TRPC3 homotetramer, this can be coordinated by at least 20 sites for lipid modification, and 16 sites for calmodulin-binding. Known PKC and PKG phosphorylation sites have been identified inclusive to 4/5 lipid-binding regions. We suggest that the negative charge of the phosphate may alter lipid-binding affinity, leading to a loss of contacts of the signal antennae with the membrane. Indeed, both PKC/PKG phosphorylation of TRPC3 inhibits channel activity.Citation13,Citation16 Finally, we propose that high calcium induces multiple calmodulin/TRPC3 interactions. This complex may underlie the deactivation of the channel by shielding the signaling antennae from protein and/or lipid contacts. This idea is supported by the findings that TRPC channels bind to calmodulin via their C-termini as well as additional regions in the N-terminus.
Conclusions
Our functional models of TRPC3 are far from “high-definition”. For instance, we have not yet modeled other known regulatory elements for these channels including homer, IP3R and nitrosylation. Further, our current models cannot predict, de novo, the functional consequences of such interactions (e.g., what does calmodulin do when bound to the N-terminus? Which PIP2-binding sites are stimulatory vs. inhibitory?). Added complexity stems from the ability of TRPC channels to form homo- and heteromultimers; our models cannot predict how these domains control function within a specific tetramer. That being stated, even in this nascent stage, GDDA-BLAST analysis can speed the discovery process for the bench scientist by providing a rationale for directed experimentation including mutagenesis. Results from such experiments can be iterated back onto our computational models, towards improving the resolution of our computational measures. In closing, we propose that additional development of GDDA-BLAST will lead to refined phylogenetic profiles which can accurately predict protein domain function as well as the specificity of their interactions.
Figures and Tables
Figure 1 Structure/Function Modeling of TRPC3. (A) GDDA-BLAST histogram depicting phylogenetic profile signals for peripheral lipid-binding (magenta, 131 profiles), integral lipid-binding (yellow, 98 profiles), ankyrin repeats (turquoise, 54 profiles) and calmodulin-binding (green, 166 profiles). We observe multiple calmodulin-binding domains that overlap with lipid-binding domains in the N-terminal ankyrin repeats, as well as a major site in the C-terminus. These results predict at least 5 lipid-binding domains and 4 calmodulin-binding sites in TRPC3. (Functional phylogenetic profiles for TRP channels available upon request) (B) left: Cryo EM structure of TRPC3 from Mio et al.Citation3 where they suggest the N-terminal and C-terminal tails of TRPC3 form signal-sensing antennae. right: Artistic representation of the domain architecture for TRPC3. The N-terminal and C-terminal regions, channel, inner-shell and ankyrin repeats are approximated from the Mio et al. study.Citation3 The other regions are based on the overall topology when compared with our GDDA-BLAST results. Based on this model, each region of the channel predicted to interact with lipid/calmodulin has a solvent-accessible surface that would allow for protein/lipid-interactions, particularly if these regions moved to contact the plasma membrane.
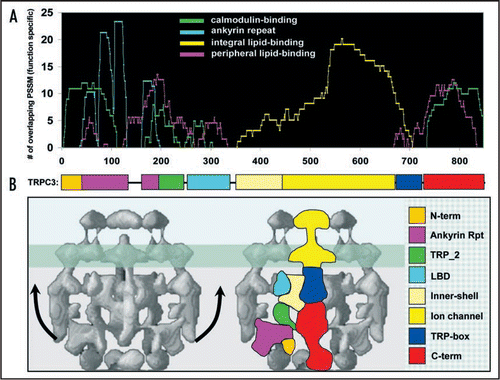
Figure 2 Theoretical Model for the TRPC3 life cycle. Components: (left) Graphical representation of the lipid-binding (magenta) and calmodulin-binding (green) domain predictions from GDDA-BLAST. Inset within the lipid-binding domains, lipid-binding specificity of each region is depicted. Below these predictions are known and predicted (S209/S213) PKC and PKG phosphorylation sites (red),Citation15,Citation16 and the binding regions for known TRPC3-interacting proteins.Citation5,Citation6,Citation8,Citation9,Citation12,Citation25–Citation27 We observe that the phosphorylation and protein interactions sites correlate with our lipid/calmodulin interaction sites, suggesting multiple regulation mechanisms for these interactions, which has been observed for TRP channels.Citation8 (right) Graphical representation of the lipid-binding, calmodulin binding and phosphorylation sites for a TRPC3 homo-tetramer. (PL = phospholipids, S1P = sphingosine-1-phosphate) Insertion: Receptor activation (GPCR, RTK, etc.,) signals for the activation of PLC, generating DAG and IP3, as well as signaling for VAMP2-dependent trafficking of TRPC3 to the plasma membrane, where the vesicle docks.Citation5 Inset: Docking occurs through VAMP2 interactions with SNARE complexes, as well as TRPC3 TRP_2 lipid-binding.Citation4 Following, increased DAG levels destabilize the plasma membrane, alleviating TRPC3 TRP_2 domain lipid-binding, facilitating vesicle fusion.Citation4 Activation: We propose that once inserted, the TRP_2 domain associates with phospholipids, stabilizing the ion-channel in the membrane. Following, PLCγ binds to the N-terminus of TRPC3, where the channel can then “search” for PIP2.Citation9 Concomitantly, the C-terminus of TRPC3 can search for PIP3,Citation11 spatially localizing the channel, as has been observed for other membrane-associated proteins such as PhopholipaseA2.Citation28 Once properly localized, interactions with both lipids and proteins (IP3R, RACK1, Homer, PLCγ) control the overall activity of the channel. We propose that these interactions swell the ion-channel by placing the N- and C-terminal tails of TRPC3 parallel to the plasma membrane. Deactivation: Increased Ca2+ and DAG levels allow PKC to phosphorylate TRPC3 within lipid/protein binding domains, altering these interactions.Citation8 Increased Ca2+ also activates calmodulin, which associates with phosphorylated TRPC3, blocking the protein/lipid-interactions. Losses of these interactions allow the N- and C-terminus of TRPC3 to dissociate from the plasma membrane, closing the channel. Following, the channel can be endocytosed through a Homer-dependent pathway.Citation26 Upon endocytosis and decreases in intracellular Ca2+, calmodulin is released from TRPC3, allowing the channel to be “reset” for recycling to the plasma membrane, or targeted for degradation.
Acknowledgements
We thank Jason Holmes at the Pennsylvania State University CAC center for technical assistance; Drs. Robert E. Rothe, Jim White, B.D. Payne, B. Mandelbrot and B.H. Obama for creative dialogue. This work was supported by the Searle Young Investigators Award and start-up monies from Pennsylvania State University (Randen L. Patterson), Funds from the Huck Life Science Institute's Center for Computational Proteomics (Randen L. Patterson and Damian B. van Rossum) and a grant from the Pennsylvania Department of Health using Tobacco Settlement Funds to Damian B. van Rossum. The Pennsylvania Department of Health specifically disclaims responsibility for any analyses, interpretations or conclusions.
Addendum to:
References
- Ko KD, Hong Y, Chang GS, Bhardwaj G, van Rossum DB, Patterson RL. Physics Archives 2008; arXiv:0806.239, q-bio.Q
- Chang GS, Hong Y, Ko KD, Bhardwaj G, Holmes EC, Patterson RL, et al. Phylogenetic profiles reveal evolutionary relationships within the “twilight zone” of sequence similarity. Proc Natl Acad Sci USA 2008; 105:13474 - 13479
- Mio K, Ogura T, Kiyonaka S, Hiroaki Y, Tanimura Y, Fujiyoshi Y, et al. The TRPC3 channel has a large internal chamber surrounded by signal sensing antennas. J Mol Biol 2007; 367:373 - 383
- van Rossum DB, Oberdick D, Rbaibi Y, Bhardwaj G, Barrow RK, Nikolaidis N, et al. TRP_2, a lipid/trafficking domain that mediates diacylglycerol-induced vesicle fusion. J Biol Chem 2008; 283:34384 - 34392
- Singh BB, Lockwich TP, Bandyopadhyay BC, Liu X, Bollimuntha S, Brazer SC, et al. VAMP2-dependent exocytosis regulates plasma membrane insertion of TRPC3 channels and contributes to agonist-stimulated Ca2+ influx. Mol Cell 2004; 15:635 - 646
- Bandyopadhyay BC, Ong HL, Lockwich TP, Liu X, Paria BC, Singh BB, et al. TRPC3 controls agonist-stimulated intracellular Ca2+ release by mediating the interaction between inositol 1,4,5-trisphosphate receptor and RACK1. J Biol Chem 2008; 283:32821 - 32830
- Kiselyov KI, Xu X, Mohayeva G, Kuo T, Pessah IN, Mignery GA, et al. Functional interaction between InsP3 receptors and store-operated Htrp3 channels. Nature 1998; 396:478 - 482
- Zhu MX. Multiple roles of calmodulin and other Ca(2+)-binding proteins in the functional regulation of TRP channels. Pflugers Arch 2005; 451:105 - 115
- van Rossum DB, Patterson RL, Sharma S, Barrow RK, Kornberg M, Gill DL, et al. Phospholipase Cgamma1 controls surface expression of TRPC3 through an intermolecular PH domain. Nature 2005; 434:99 - 104
- Lemonnier L, Trebak M, Putney JW Jr. Complex regulation of the TRPC3, 6 and 7 channel subfamily by diacylglycerol and phosphatidylinositol-4,5-bisphosphate. Cell Calcium 2008; 43:506 - 514
- Kwon Y, Hofmann T, Montell C. Integration of phosphoinositide- and calmodulin-mediated regulation of TRPC6. Mol Cell 2007; 25:491 - 503
- Boulay G, Brown DM, Qin N, Jiang M, Dietrich A, Zhu MX, et al. Modulation of Ca(2+) entry by polypeptides of the inositol 1,4,5-trisphosphate receptor (IP3R) that bind transient receptor potential (TRP): evidence for roles of TRP and IP3R in store depletion-activated Ca(2+) entry. Proc Natl Acad Sci USA 1999; 96:14955 - 14960
- Venkatachalam K, Zheng F, Gill DL. Regulation of canonical transient receptor potential (TRPC) channel function by diacylglycerol and protein kinase C. J Biol Chem 2003; 278:29031 - 29040
- Yoshida T, Inoue R, Morii T, Takahashi N, Yamamoto S, Hara Y, et al. Nitric oxide activates TRP channels by cysteine S-nitrosylation. Nat Chem Biol 2006; 2:596 - 607
- Kim JY, Saffen D. Activation of M1 muscarinic acetylcholine receptors stimulates the formation of a multiprotein complex centered on TRPC6 channels. J Biol Chem 2005; 280:32035 - 32047
- Kwan HY, Huang Y, Yao X. Protein kinase C can inhibit TRPC3 channels indirectly via stimulating protein kinase G. J Cell Physiol 2006; 207:315 - 321
- Kwan HY, Huang Y, Yao X. Regulation of canonical transient receptor potential isoform 3 (TRPC3) channel by protein kinase G. Proc Natl Acad Sci USA 2004; 101:2625 - 2630
- van Rossum DB, Oberdick D, Rbaibi Y, Bhardwaj G, Barrow RK, Snyder SH, et al. Phospholipase Cgamma1 controls surface expression of TRPC3 through an intermolecular PH domain. J Biol Chem 2008; 283:34384 - 34392
- Nilius B, Mahieu F, Prenen J, Janssens A, Owsianik G, Vennekens R, et al. The Ca2+-activated cation channel TRPM4 is regulated by phosphatidylinositol 4,5-biphosphate. EMBO J 2006; 25:467 - 478
- Krapivinsky G, Mochida S, Krapivinsky L, Cibulsky SM, Clapham DE. The TRPM7 ion channel functions in cholinergic synaptic vesicles and affects transmitter release. Neuron 2006; 52:485 - 496
- Hofmann T, Obukhov AG, Schaefer M, Harteneck C, Gudermann T, Schultz G. Direct activation of human TRPC6 and TRPC3 channels by diacylglycerol. Nature 1999; 397:259 - 263
- Trebak M, St J Bird G, McKay RR, Birnbaumer L, Putney JW Jr. Signaling mechanism for receptor-activated canonical transient receptor potential 3 (TRPC3) channels. J Biol Chem 2003; 278:16244 - 16252
- Mio K, Ogura T, Hara Y, Mori Y, Sato C. The non-selective cation-permeable channel TRPC3 is a tetrahedron with a cap on the large cytoplasmic end. Biochem Biophys Res Commun 2005; 333:768 - 777
- Kiselyov KI, Mignery GA, Zhu MX, Muallem S. The N-terminal domain of the IP3 receptor gates store-operated hTrp3 channels. Mol Cell 1999; 4:423 - 429
- Yuan JP, Kiselyov KI, Shin DM, Chen J, Shcheynikov N, Kang HS, et al. Homer binds TRPC family channels and is required for gating of TRPC1 by IP3 receptors. Cell 2003; 114:777 - 789
- Kim JY, Zeng W, Kiselyov K, Yuan JP, Dehoff MH, Mikoshiba K, et al. Homer 1 mediates store- and inositol 1,4,5-trisphosphate receptor-dependent translocation and retrieval of TRPC3 to the plasma membrane. J Biol Chem 2006; 281:32540 - 32549
- Caraveo G, van Rossum DB, Patterson RL, Snyder SH, Desiderio S. Action of TFII-I outside the nucleus as an inhibitor of agonist-induced calcium entry. Science 2006; 314:122 - 125
- Winget JM, Pan YH, Bahnson BJ. The interfacial binding surface of phospholipase A2s. Biochim Biophys Acta 2006; 1761:1260 - 1269