Abstract
The Retinoblastoma (pRb) pathway has been implicated as a convergent regulatory unit in the control of cell cycle and disease. We have shown that a crosstalk between RETINOBLASTOMA RELATED (RBR), the Arabidopsis homologue of pRb, and the genes encoding proteins of the chromatin complexes involved in DNA or histone methylation, controls gametophytic and post-fertilization differentiation events and a subset of imprinting effects. We describe here a plausible model that incorporates several components of the plant Retinoblastoma pathway, thus offering a novel paradigm that merges the traditional cell cycle and the chromatin components in the control of cell differentiation and imprinting.
The short-lived male and female gametophytes are the two haploid phases of the plant life cycle that are independently derived from the dominant diploid sporophyte as a consequence of meiosis.Citation1 Gametophytes produce gametes and accessory cell types through a series of nuclear divisions, cell specification and differentiation events. At maturity, plant gametes are represented by an egg and a central cell in the female gametophyte, and two sperm cells in the male gametophyte (). Fertilization of an egg by a sperm cell marks the completion of the gametophytic phase. It constitutes the maternal to embryonic transition by forming a diploid embryo, the sporophyte. At the same time the central cell is fertilized by a second sperm in all angiosperms. This gives rise to the endosperm, which is an extra-embryonic annex that nourishes the embryo. Both the gametophytic (central cell and sperm cell) and endosperm developmental phases are essential reproductive platforms in plants, in which imprinting effects that epigenetically distinguish parent-of-origin specific gene expression are established and/or maintained primarily by DNA and/or histone methylation.Citation2
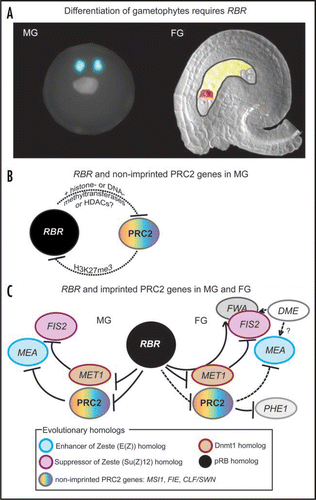
ReTINoBLAsToMA ReLATeD (RBR) is a single copy plant homologue of Retinoblastoma (pRb), a key cell cycle regulatory protein and also a tumor suppressor in animals.Citation3 In our recent work, we have shown that in Arabidopsis thaliana RBR is essential for appropriate differentiation of all gametic and accessory cell types of the gametophytes ().Citation4 In our attempt to identify convergent mechanisms controlling cell differentiation, we observed that loss of RBR derepresses several genes encoding proteins of the chromatin-associated Polycomb Repressive Complex 2 (PRC2) in both gametophytes. For example, an Arabidopsis PRC2 histone methyltransferase, CURLY LeAF,Citation5 which is a homologue of the Drosophila gene enhancer of zeste [E(z)] that has a canonical E2F binding motif in its promoter and a predicted RBR binding motif (LXCXD) within its protein sequence, was strongly upregulated in rbr male and female gametophytes. Similarly, the Arabidopsis METHYL TRANSFERASE 1 (MET1), an evolutionary homologue of the mammalian gene DNA Methyltransferase 1 (Dnmt1)Citation6 and a direct target of the RBR/E2F/MSI1 (RbAP46/48) complex,Citation4,Citation7 is derepressed in rbr gametophytic cells. It must be noted that in the both cases described above, similar transcriptional repression was demonstrated in animal cells for the convergent homologues of E(z) and Dnmt1,Citation8,Citation9 however, if similar mechanism operates in animal gametes is unknown. Considering that the evolutionary homologues of pRb, E(z) and Dnmt1 play critical roles in cell differentiation,Citation9,Citation10 our results reiterate the synergism between the cell cycle and members of the chromatin-bound repressive complexes in gene silencing and cellular differentiation.
The interplay of key cell cycle regulators and chromatin associated proteins is expected to control the epigenetic differentiation of the endosperm from the fertilized central cells. Precocious activation of cell divisions in central cells is prevented until fertilization occurs, and presumably the chromatin structure is altered as part of the epigenetic reprogramming.Citation4,Citation11 Considering that rbr central cells initiate endosperm-like proliferation and heterochromatin status in the absence of fertilization similar to the maternal-effect mutants of the MEDEA (MEA)-PRC2 Fertilization-Independent-Seed (FIS) complex,Citation2 we asked if the PRC2 members would cross-talk with RBR. Although we were unable to examine how RBR is regulated in the female tissues of the maternal fis mutants due to technical limitations, we observed that the maternal PRC2, and most likely a paternal PRC2 as well, target the paternal RBR allele in endosperm or pollen tissues. This particular finding of a regulatory circuit between RBR and PRC2 is novel and interesting. In mammals, cell cycle exit is controlled by pRb partly by regulating the PRC2 specific histone mark H3K27me3 on cell cycle genes.Citation12 However, it remains to be demonstrated if the PRC2 and/or similar chromatin complexes would directly control pRb as well. It will therefore be intriguing to understand if parent-of-origin dependent gene targeting of pRb has operated in evolution.
Imprinting is a fascinating epigenetic process that evolved in mammals and plants for their placental behaviour.Citation2 Although imprinting mechanisms are conserved in evolution, expression of sex-specific alleles in plants is established by differential erasure of silencing marks during gametophyte development. silencing marks can vary from DNA methylation, which is maintained by MET1 in the case of paternally imprinted FIS2 or FLOWERING LOCUS A (FWA), to histone methylation in case of MEA, or both DNA and histone methylation for PHERES1 (PHE1).Citation2,Citation13 It has also been shown that demethylation in central cells is achieved by a base-excision DNA repair mechanism involving DNA glycosylase, DEMETER (DME).Citation2 DME erases the silencing marks from FIS2 and FWA locus by removing methylated cytosines, thus activating the corresponding maternal alleles in the central cells. While we could not obtain convincing evidence for the control of DME by RBR, our recent findingsCitation4 and the work of Julliene and co-workersCitation7 suggest that erasure of the maternal FIS2 (and FWA) imprint mediated by MET1 could be directly controlled by a RBR (and MSI1) complex via RBR/E2F mediated direct repression of S-phase specific MET1 expression during each G1-phase. Therefore, we propose that repression of MET1 activity by the RBR pathway likely initiates global DNA demethylation during gametogenesis, which is required for the activation of a subset of imprinted genes. While we are currently developing conditional rbr mutants to understand how RBR regulates PRC2, MET1 and ultimately the imprinting effects during endosperm development, our working hypothesis predicts that the maternal activity of PRC2 suppresses RBR in the endosperm tissues, whereas MET1-dependent expression of the imprinted FIS2, FWA and likely PHERES1 genes is maintained in a dynamic fashion. Together, our work provides the first insight that the evolutionarily ancient RBR, PRC2 and MET1 networks could predate the separation of animals and plants, and their regulatory interaction would play a significant role in mediating certain imprinting effects during gametic and post-fertilization differentiation events.
Abbreviations
| RBR | = | retinoblastoma related |
| PRC2 | = | polycomb repressive complex 2 |
| MET1 | = | DNA methyltransferase 1 |
| FIS | = | fertilization-independent-seed |
Figures and Tables
Figure 1 Epigenetic interaction between RBR and chromatin-associated regulatory proteins and complexes is critical for gametophyte development. (A) Sketch of fully differentiated Arabidopsis male (MG) and female (FG) gametophytes in the presence of RBR. Gametophytic cells such as sperm, egg and central cells are marked in blue, red and yellow, respectively. (B and C) Models illustrating the regulatory interactions of RBR, MET1, PRC2 and its targets. For simplicity, only the mature male (MG) and female gametophyte (MG) specific gene regulation prior to fertilization is shown. Post-fertilization interaction between RBR and the PRC2 genes in the embryo and endosperm could likely be similar.
Acknowledgements
The research was funded by the swiss National science Foundation grant to Wilhelm Gruissemm. Amal J. Johnston was supported by a post-doctoral fellowship from the Roche Research Foundation. We thank olga Kirioukhova for help with the figure and comments on the manuscript.
Addendum to:
References
- Boavida LC, Becker JD, Feijo JA. The making of gametes in higher plants. Int J Dev Biol 2005; 49:595 - 614
- Kohler C, Makarevich G. Epigenetic mechanisms governing seed development in plants. EMBO Rep 2006; 7:1223 - 1227
- Gruissem W. Inze D. Function of the Retinoblastoma-related protein in plants. Cell Cycle Control and Plant Development 2007; Oxford, UK Blackwell Publishing 164 - 186
- Johnston AJ, Matveeva E, Kirioukhova O, Grossniklaus U, Gruissem W. A dynamic reciprocal RBR-PRC2 regulatory circuit controls Arabidopsis gametophyte development. Curr Biol 2008; 18:1680 - 1686
- Goodrich J, Puangsomlee P, Martin M, Long D, Meyerowitz EM, Coupland G. A Polycomb-group gene regulates homeotic gene expression in Arabidopsis. Nature 1997; 386:44 - 51
- Finnegan EJ, Peacock WJ, Dennis ES. Reduced DNA methylation in Arabidopsis thaliana results in abnormal plant development. Proc Natl Acad Sci USA 1996; 93:8449 - 8454
- Jullien PE, Mosquna A, Ingouff M, Sakata T, Ohad N, Berger F. Retinoblastoma and its binding partner MSI1 control imprinting in Arabidopsis. PLos Biol 2008; 6:194
- Bracken AP, Pasini D, Capra M, Prosperini E, Colli E, Helin K. EZH2 is downstream of the pRB-E2F pathway, essential for proliferation and amplified in cancer. EMBO J 2003; 22:5323 - 5335
- Ferreira R, Naguibneva I, Pritchard LL, Ait-Si-Ali S, Harel-Bellan A. The Rb/chromatin connection and epigenetic control: opinion. Oncogene 2001; 20:3128 - 3133
- Schwartz YB, Pirrotta V. Polycomb silencing mechanisms and the management of genomic programmes. Nat Rev Genet 2007; 8:9 - 22
- Baroux C, Pecinka A, Fuchs J, Schubert I, Grossniklaus U. The triploid endosperm genome of Arabidopsis adopts a peculiar, parental-dosage-dependent chromatin organization. Plant Cell 2007; 19:1782 - 1794
- Blais A, van Oevelen CJ, Margueron R, Acosta-Alvear D, Dynlacht BD. Retinoblastoma tumor suppressor protein-dependent methylation of histone H3 lysine 27 is associated with irreversible cell cycle exit. J Cell Biol 2007; 179:1399 - 1412
- Makarevich G, Villar CB, erilova A, Kohler C. Mechanism of PHERES1 imprinting in Arabidopsis. J Cell sci 2008; 121:906 - 912
- McCabe MT, Davis JN, Day ML. Regulation of DNA methyltransferase 1 by the pRb/E2F1 pathway. Cancer Res 2005; 65:3624 - 3632