Abstract
Mitochondrial metabolism has traditionally been thought of as a source of cellular energy in the form of ATP. The recent renaissance in the study of cellular metabolism, particularly in the cancer field, has highlighted the fact that mitochondria are also critical biosynthetic and signaling hubs, making these organelles key governors of cellular outcomes.Citation1-Citation5 Using the epidermis as a model system, our recent study looked into the role that mitochondrial metabolism and ROS production play in cellular differentiation in vivo.Citation6 We showed that conditional deletion of the mitochondrial transcription factor, TFAM within the basal cells of the epidermis results in loss of mitochondrial ROS production and impairs epidermal differentiation and hair growth. We demonstrated that mitochondrial ROS generation is required for the propagation of Notch and β-catenin signals which promote epidermal differentiation and hair follicle development respectively. This study bolsters accumulating evidence that oxidative mitochondrial metabolism plays a causal role in cellular differentiation programs. It also provides insights into the role that mitochondrial oxidative signaling plays in a cell type-dependent manner.
Mitochondrial ROS Regulate Signal Transduction
Mitochondria produce reactive oxygen species (ROS) during oxidative metabolism through the single-electron reduction of molecular oxygen (O2).Citation7 The resulting superoxide anion (O2•−), is converted to hydrogen peroxide (H2O2) by cellular superoxide dismutases (SODs). Similar to phosphorylation, ubiquitination, and acetylation, oxidation of protein cysteine residues represents a form of reversible, posttranslational protein modification which can regulate signaling. Oxidation of cysteine thiol groups (–SH) by H2O2 results in formation of sulphenic acid (–SOH) which quickly forms disulfide (–SS–) or sulfenyl amide (–SN–) bonds, changing protein function.Citation8,Citation9 The cellular thioredoxin and glutathione reductase systems restore these oxidatively modified residues back to their reduced forms in a manner analogous to phosphatases, deubiquitinating enzymes, or histone deacetylases.
Mitochondrial ROS (mROS) generation is required for the propagation of numerous cellular signaling pathways including those regulating tumorgenesis,Citation10-Citation12 immune responses,Citation13-Citation15 and cellular adaptation to stresses such as hypoxia.Citation16-Citation19 In most cases it is unknown exactly what oxidative events are required for the effects of ROS on these pathways and it is likely that multiple oxidative targets potentiate transduction through a given pathway.
Increasing evidence suggests that mitochondria and mROS generation play key roles in cellular differentiation programs. Cellular mitochondrial content and oxidative capacity are increased when mouse embryonic stem cells (ESCs), induced pluripotent stem cells (iPSCs), or mesenchymal stem cells are induced to differentiate.Citation20-Citation24 Furthermore, increased cellular ROS levels and oxidation products correlate with the differentiation of ESCs and iPSCs as well as mesenchymal, neural, and epithelial stem cells.Citation23,Citation25-Citation30 Hematopoietic stem cells (HSCs) contain significantly lower levels of ROS than do the more-differentiated common myeloid progenitor cells. Aberrantly increased ROS levels promote HSC differentiation while inhibition of ROS production prevents differentiation.Citation31-Citation34 Thus increased mitochondrial and oxidant content appears to correlate with stem cell differentiation while low mitochondrial mass and oxidant content correlates with stem cell maintenance. In our recent report, we used the mammalian epidermis as a model system to test the hypothesis that mitochondrial metabolism and ROS production promote signaling through pathways required for differentiation of mammalian stem cell populations in vivo.Citation6
Mitochondrial ROS Promote Epidermal Differentiation and Hair Growth
The epidermis is a self-renewing stratified squamous epithelium and is thus regulated by stem cell populations. To maintain epidermal homeostasis, cells within the proliferative basal layer withdraw from the cell cycle and differentiate as they process outward through the suprabasal layers, compensating for cellular loss via desquamanation from the outermost epidermal layer.Citation35 The epidermis also elaborates appendages such as hair follicles (HFs) which are themselves regulated by stem cell populations.Citation36 Multiple transcriptional networks are associated with differentiation within the epidermis including Notch, p63, C/EBP, and AP2 (interfollicular epidermis) and β-catenin (HF).Citation37 It is incompletely understood how these various factors are regulated to promote epidermal homeostasis and what role cellular metabolism might play in this regulation.
To test the hypothesis that mitochondria play an active role in the regulation of epidermal homeostasis, we conditionally deleted TFAM (transcription factor A, mitochondrial) in basal, undifferentiated epidermal keratinocytes. TFAM is required for the replication and transcription of the mitochondrial genome, and cells lacking TFAM are unable to conduct oxidative phosphorylation or produce mROS.Citation38 Mice conditionally lacking TFAM in epidermis (TFAM cKO mice) lacked hair and developed an epidermal barrier defect which contributed to their perinatal mortality. Histologically, the skin of these mice displayed signs of impaired keratinocyte differentiation.
Similar to in vivo, primary keratinocytes derived from TFAM cKO mice displayed impaired differentiation in vitro. Differentiation of wild-type keratinocytes was inhibited by antioxidant treatment, and differentiation marker expression in TFAM cKO cells could be partly restored by treatment with exogenously applied H2O2, clearly demonstrating that oxidative signaling promotes keratinocyte differentiation. We went on to demonstrate that mROS generation is required for activation of the Notch and β-catenin transcriptional programs that promote epidermal differentiation and hair growth respectively. Together, our results show that mROS act as pro-differentiation signals and are key upstream regulators of stem cell fate decisions.
While the role for mROS in signal transduction is becoming increasingly accepted, the identities of the exact targets of oxidation which promote signaling remain unknown in most situations. Our work identified nucleoredoxin (NXN) as a putative target of mROS-mediated Wnt-β-catenin signaling. NXN is a member of the thioredoxin family which has previously been demonstrated to regulate β-catenin-dependent transcription in a redox-sensitive manner.Citation39 Our results show that NXN becomes oxidized when wild-type keratinocytes are treated with the β-catenin activator Wnt-3a. This did not occur in TFAM cKO keratinocytes, suggesting that mROS-mediated NXN oxidation plays a causal role in the transduction of signals between Wnt receptors and β-catenin. Further studies will be required to determine how mROS regulate Notch transcriptional activity during keratinocyte differentiation.
Future Directions: Metabolism as the Gatekeeper to Cellular Differentiation
Our study demonstrates that mitochondrial metabolism and ROS generation promote keratinocyte differentiation. This raises the interesting possibility that aberrant keratinocyte metabolism may be associated with, and presents targets for, the treatment of epidermal disease. While the genetic causes of these diseases are increasingly studied, little is known about how disease-causing mutations might affect cellular metabolism. More generally, the interactions between cellular differentiation programs and cellular metabolism will be increasingly studied in years to come. The development of regenerative therapies depends on the ability to maximize the renewal capacity of stem cells, and subsequently, to promote complete differentiation into desired cell types. Cellular metabolism represents an intriguing “rheostat” which may be used to modulate these ends. A further understanding of the mechanisms by which differentiation programs both regulate, and are regulated by metabolism will prove useful in this regard.
It is unknown how stem and differentiated cells maintain their respective metabolic phenotypes. Proper regulation is likely dependent on cell type-specific factors as well as environmental influences such as hypoxia. In keratinocytes, elevation of extracellular calcium is the most common method for inducing differentiation in vitro. As an epidermal calcium gradient exists in vivo, it is hypothesized that calcium concentrations regulate epidermal differentiation in vivo as well.Citation40 Calcium uptake into mitochondria is known to stimulate the activity of TCA cycle enzymes (pyruvate-, isocitrate-, and oxoglutarate-dehydrogenases), promoting mitochondrial respiration.Citation41 We found that inhibition of mitochondrial calcium uptake prevented keratinocyte differentiation in vitro, suggesting that mitochondrial calcium uptake might promote the mitochondrial metabolism and ROS production required for epidermal differentiation.
A recent study suggests that mitochondrial uncoupling proteins may play a role in regulating differentiation-dependent metabolism.Citation42 Uncoupling protein 2 (UCP2) is highly expressed in iPSCs, with reduced expression in differentiated cells. Ectopic UCP2 expression inhibited oxidative metabolism of pyruvate, reduced cellular ROS content, and prevented differentiation suggesting that this protein could act as a metabolic switch that controls stem cell differentiation. How UCP2 expression is repressed during differentiation remains to be determined and it will also be of interest to determine if UCP2 regulates the metabolism of less-pluripotent, tissue-specific stem cell populations.
Beyond promotion of oxidant-dependent signaling, the mechanisms by which mitochondrial metabolism promotes cellular differentiation remain poorly understood. Some differentiated lineages may require the efficient ATP production provided by oxidative phosphorylation; however, another intriguing possibility involves the nutrient-dependent regulation of chromatin (). Acetyl-CoA, a key fuel for oxidative metabolism, is also a substrate used by histone acetyltransferases to acetylate histones and other cellular proteins. Global increases in histone acetylation are associated with differentiation of ESCs, 3T3-L1 preadipocytes, and keratinocytes.Citation43-Citation45 Treatment of primary human keratinocytes with histone deacetylase inhibitors is sufficient to induce cell cycle arrest and expression of several differentiation markers.Citation46,Citation47 Although differentiation programs likely regulate histone acetylation in a gene and time-dependent manner; these reports suggest that cellular differentiation may depend on availability of acetyl-CoA to carry out acetylation reactions. In mammalian cells, most cytosolic acetyl-CoA is produced through the cleavage of TCA-cycle-derived citrate by the enzyme ATP-citrate lyase (ACL). Inhibition of ACL expression prevents global increases in histone acetylation and differentiation of 3T3-L1 cells.Citation48
Figure 1. Mitochondrial metabolism promotes keratinocyte differentiation. During oxidative metabolism, electrons are transferred from TCA cycle substrates to complexes I and II of the electron transport chain via NADH and FADH2 respectively. These electrons are transferred to complex III (via coenzyme Q) and subsequently to complex IV (via cytochrome c) where they are used to reduce molecular oxygen to water. Complexes I, II, and III of the respiratory chain contain sites in which electrons can prematurely react with oxygen, producing ROS. Our recent report demonstrates that mitochondrial ROS production is required for activation of Notch and β-catenin transcriptional programs which promote epidermal differentiation and hair development respectively. Mitochondrial metabolism may also promote keratinocyte differentiation by providing substrates for histone acetyltransferases (HATs) and histone demethylases (such as JMJD3).
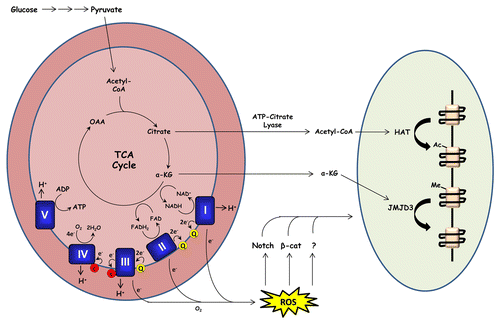
Another histone modification potentially regulated by cellular metabolism is methylation. Histone methylation status is regulated by histone methyltransferases and demethylases with various methylation “codes” associated with either transcriptional activation or repression. One particular methyl modification—trimethylation of histone 3 on lysine 27 (H3K27me3)—is associated with transcriptional repression of epidermal differentiation markers.Citation49 Genes such as keratin 1 and involucrin display high levels of H3K27me3 in undifferentiated keratinocytes, and lose this mark as cells differentiate. Demethylation of H3K27me3 requires the demethylase JMJD3 and cells lacking JMJD3 expression are unable to differentiate in culture.Citation49 JMJD3 is a member of the Jumanji-C domain containing family of demethylases which use oxygen, Fe(II), and the TCA cycle intermediate α-ketoglutarate as cofactors and are thus potentially sensitive to cellular metabolic state.Citation50,Citation51
Many other nutrient-sensitive mechanisms are likely to regulate stem cell maintenance and differentiation in both the epidermis and in other compartments. Cancer researchers are currently striving to identify cancer-specific metabolic signatures which can be therapeutically targeted. Metabolic study of cellular differentiation is an emerging area which, in concert with genetic approaches, has the potential to lead to strategies for targeted control of cell fate decisions and therapies for diseases associated with aging.
References
- DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab 2008; 7:11 - 20; http://dx.doi.org/10.1016/j.cmet.2007.10.002; PMID: 18177721
- Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 2009; 324:1029 - 33; http://dx.doi.org/10.1126/science.1160809; PMID: 19460998
- Cairns RA, Harris IS, Mak TW. Regulation of cancer cell metabolism. Nat Rev Cancer 2011; 11:85 - 95; http://dx.doi.org/10.1038/nrc2981; PMID: 21258394
- Folmes CD, Dzeja PP, Nelson TJ, Terzic A. Metabolic plasticity in stem cell homeostasis and differentiation. Cell Stem Cell 2012; 11:596 - 606; http://dx.doi.org/10.1016/j.stem.2012.10.002; PMID: 23122287
- Hamanaka RB, Chandel NS. Mitochondrial reactive oxygen species regulate cellular signaling and dictate biological outcomes. Trends Biochem Sci 2010; 35:505 - 13; http://dx.doi.org/10.1016/j.tibs.2010.04.002; PMID: 20430626
- Hamanaka RB, Glasauer A, Hoover P, Yang S, Blatt H, Mullen AR, et al. Mitochondrial reactive oxygen species promote epidermal differentiation and hair follicle development. Sci Signal 2013; 6:ra8; http://dx.doi.org/10.1126/scisignal.2003638; PMID: 23386745
- Turrens JF. Mitochondrial formation of reactive oxygen species. J Physiol 2003; 552:335 - 44; http://dx.doi.org/10.1113/jphysiol.2003.049478; PMID: 14561818
- Janssen-Heininger YM, Mossman BT, Heintz NH, Forman HJ, Kalyanaraman B, Finkel T, et al. Redox-based regulation of signal transduction: principles, pitfalls, and promises. Free Radic Biol Med 2008; 45:1 - 17; http://dx.doi.org/10.1016/j.freeradbiomed.2008.03.011; PMID: 18423411
- Brandes N, Schmitt S, Jakob U. Thiol-based redox switches in eukaryotic proteins. Antioxid Redox Signal 2009; 11:997 - 1014; http://dx.doi.org/10.1089/ars.2008.2285; PMID: 18999917
- Weinberg F, Hamanaka R, Wheaton WW, Weinberg S, Joseph J, Lopez M, et al. Mitochondrial metabolism and ROS generation are essential for Kras-mediated tumorigenicity. Proc Natl Acad Sci U S A 2010; 107:8788 - 93; http://dx.doi.org/10.1073/pnas.1003428107; PMID: 20421486
- Li T, Kon N, Jiang L, Tan M, Ludwig T, Zhao Y, et al. Tumor suppression in the absence of p53-mediated cell-cycle arrest, apoptosis, and senescence. Cell 2012; 149:1269 - 83; http://dx.doi.org/10.1016/j.cell.2012.04.026; PMID: 22682249
- Kim HS, Patel K, Muldoon-Jacobs K, Bisht KS, Aykin-Burns N, Pennington JD, et al. SIRT3 is a mitochondria-localized tumor suppressor required for maintenance of mitochondrial integrity and metabolism during stress. Cancer Cell 2010; 17:41 - 52; http://dx.doi.org/10.1016/j.ccr.2009.11.023; PMID: 20129246
- Zhou R, Yazdi AS, Menu P, Tschopp J. A role for mitochondria in NLRP3 inflammasome activation. Nature 2011; 469:221 - 5; http://dx.doi.org/10.1038/nature09663; PMID: 21124315
- Nakahira K, Haspel JA, Rathinam VA, Lee SJ, Dolinay T, Lam HC, et al. Autophagy proteins regulate innate immune responses by inhibiting the release of mitochondrial DNA mediated by the NALP3 inflammasome. Nat Immunol 2011; 12:222 - 30; http://dx.doi.org/10.1038/ni.1980; PMID: 21151103
- Sena LA, Li S, Jairaman A, Prakriya M, Ezponda T, Hildeman DA, et al. Mitochondria are required for antigen-specific T cell activation through reactive oxygen species signaling. Immunity 2013; 38:225 - 36; http://dx.doi.org/10.1016/j.immuni.2012.10.020; PMID: 23415911
- Chandel NS, Maltepe E, Goldwasser E, Mathieu CE, Simon MC, Schumacker PT. Mitochondrial reactive oxygen species trigger hypoxia-induced transcription. Proc Natl Acad Sci U S A 1998; 95:11715 - 20; http://dx.doi.org/10.1073/pnas.95.20.11715; PMID: 9751731
- Brunelle JK, Bell EL, Quesada NM, Vercauteren K, Tiranti V, Zeviani M, et al. Oxygen sensing requires mitochondrial ROS but not oxidative phosphorylation. Cell Metab 2005; 1:409 - 14; http://dx.doi.org/10.1016/j.cmet.2005.05.002; PMID: 16054090
- Guzy RD, Hoyos B, Robin E, Chen H, Liu L, Mansfield KD, et al. Mitochondrial complex III is required for hypoxia-induced ROS production and cellular oxygen sensing. Cell Metab 2005; 1:401 - 8; http://dx.doi.org/10.1016/j.cmet.2005.05.001; PMID: 16054089
- Mansfield KD, Guzy RD, Pan Y, Young RM, Cash TP, Schumacker PT, et al. Mitochondrial dysfunction resulting from loss of cytochrome c impairs cellular oxygen sensing and hypoxic HIF-alpha activation. Cell Metab 2005; 1:393 - 9; http://dx.doi.org/10.1016/j.cmet.2005.05.003; PMID: 16054088
- Facucho-Oliveira JM, Alderson J, Spikings EC, Egginton S, St John JC. Mitochondrial DNA replication during differentiation of murine embryonic stem cells. J Cell Sci 2007; 120:4025 - 34; http://dx.doi.org/10.1242/jcs.016972; PMID: 17971411
- Chung S, Arrell DK, Faustino RS, Terzic A, Dzeja PP. Glycolytic network restructuring integral to the energetics of embryonic stem cell cardiac differentiation. J Mol Cell Cardiol 2010; 48:725 - 34; http://dx.doi.org/10.1016/j.yjmcc.2009.12.014; PMID: 20045004
- Chung S, Dzeja PP, Faustino RS, Perez-Terzic C, Behfar A, Terzic A. Mitochondrial oxidative metabolism is required for the cardiac differentiation of stem cells. Nat Clin Pract Cardiovasc Med 2007; 4:Suppl 1 S60 - 7; http://dx.doi.org/10.1038/ncpcardio0766; PMID: 17230217
- Prigione A, Fauler B, Lurz R, Lehrach H, Adjaye J. The senescence-related mitochondrial/oxidative stress pathway is repressed in human induced pluripotent stem cells. Stem Cells 2010; 28:721 - 33; http://dx.doi.org/10.1002/stem.404; PMID: 20201066
- Chen CT, Shih YR, Kuo TK, Lee OK, Wei YH. Coordinated changes of mitochondrial biogenesis and antioxidant enzymes during osteogenic differentiation of human mesenchymal stem cells. Stem Cells 2008; 26:960 - 8; http://dx.doi.org/10.1634/stemcells.2007-0509; PMID: 18218821
- Yanes O, Clark J, Wong DM, Patti GJ, Sánchez-Ruiz A, Benton HP, et al. Metabolic oxidation regulates embryonic stem cell differentiation. Nat Chem Biol 2010; 6:411 - 7; http://dx.doi.org/10.1038/nchembio.364; PMID: 20436487
- Armstrong L, Tilgner K, Saretzki G, Atkinson SP, Stojkovic M, Moreno R, et al. Human induced pluripotent stem cell lines show stress defense mechanisms and mitochondrial regulation similar to those of human embryonic stem cells. Stem Cells 2010; 28:661 - 73; http://dx.doi.org/10.1002/stem.307; PMID: 20073085
- Tormos KV, Anso E, Hamanaka RB, Eisenbart J, Joseph J, Kalyanaraman B, et al. Mitochondrial complex III ROS regulate adipocyte differentiation. Cell Metab 2011; 14:537 - 44; http://dx.doi.org/10.1016/j.cmet.2011.08.007; PMID: 21982713
- Diehn M, Cho RW, Lobo NA, Kalisky T, Dorie MJ, Kulp AN, et al. Association of reactive oxygen species levels and radioresistance in cancer stem cells. Nature 2009; 458:780 - 3; http://dx.doi.org/10.1038/nature07733; PMID: 19194462
- Smith J, Ladi E, Mayer-Proschel M, Noble M. Redox state is a central modulator of the balance between self-renewal and differentiation in a dividing glial precursor cell. Proc Natl Acad Sci U S A 2000; 97:10032 - 7; http://dx.doi.org/10.1073/pnas.170209797; PMID: 10944195
- Tsatmali M, Walcott EC, Crossin KL. Newborn neurons acquire high levels of reactive oxygen species and increased mitochondrial proteins upon differentiation from progenitors. Brain Res 2005; 1040:137 - 50; http://dx.doi.org/10.1016/j.brainres.2005.01.087; PMID: 15804435
- Tothova Z, Kollipara R, Huntly BJ, Lee BH, Castrillon DH, Cullen DE, et al. FoxOs are critical mediators of hematopoietic stem cell resistance to physiologic oxidative stress. Cell 2007; 128:325 - 39; http://dx.doi.org/10.1016/j.cell.2007.01.003; PMID: 17254970
- Ito K, Hirao A, Arai F, Takubo K, Matsuoka S, Miyamoto K, et al. Reactive oxygen species act through p38 MAPK to limit the lifespan of hematopoietic stem cells. Nat Med 2006; 12:446 - 51; http://dx.doi.org/10.1038/nm1388; PMID: 16565722
- Owusu-Ansah E, Banerjee U. Reactive oxygen species prime Drosophila haematopoietic progenitors for differentiation. Nature 2009; 461:537 - 41; http://dx.doi.org/10.1038/nature08313; PMID: 19727075
- Juntilla MM, Patil VD, Calamito M, Joshi RP, Birnbaum MJ, Koretzky GA. AKT1 and AKT2 maintain hematopoietic stem cell function by regulating reactive oxygen species. Blood 2010; 115:4030 - 8; http://dx.doi.org/10.1182/blood-2009-09-241000; PMID: 20354168
- Fuchs E. Finding one’s niche in the skin. Cell Stem Cell 2009; 4:499 - 502; http://dx.doi.org/10.1016/j.stem.2009.05.001; PMID: 19497277
- Alonso L, Fuchs E. The hair cycle. J Cell Sci 2006; 119:391 - 3; http://dx.doi.org/10.1242/jcs02793; PMID: 16443746
- Blanpain C, Fuchs E. Epidermal homeostasis: a balancing act of stem cells in the skin. Nat Rev Mol Cell Biol 2009; 10:207 - 17; http://dx.doi.org/10.1038/nrm2636; PMID: 19209183
- Larsson NG, Wang J, Wilhelmsson H, Oldfors A, Rustin P, Lewandoski M, et al. Mitochondrial transcription factor A is necessary for mtDNA maintenance and embryogenesis in mice. Nat Genet 1998; 18:231 - 6; http://dx.doi.org/10.1038/ng0398-231; PMID: 9500544
- Funato Y, Michiue T, Asashima M, Miki H. The thioredoxin-related redox-regulating protein nucleoredoxin inhibits Wnt-beta-catenin signalling through dishevelled. Nat Cell Biol 2006; 8:501 - 8; http://dx.doi.org/10.1038/ncb1405; PMID: 16604061
- Dotto GP. Signal transduction pathways controlling the switch between keratinocyte growth and differentiation. Crit Rev Oral Biol Med 1999; 10:442 - 57; http://dx.doi.org/10.1177/10454411990100040201; PMID: 10634582
- Rizzuto R, Bernardi P, Pozzan T. Mitochondria as all-round players of the calcium game. J Physiol 2000; 529:37 - 47; http://dx.doi.org/10.1111/j.1469-7793.2000.00037.x; PMID: 11080249
- Zhang J, Khvorostov I, Hong JS, Oktay Y, Vergnes L, Nuebel E, et al. UCP2 regulates energy metabolism and differentiation potential of human pluripotent stem cells. EMBO J 2011; 30:4860 - 73; http://dx.doi.org/10.1038/emboj.2011.401; PMID: 22085932
- McCool KW, Xu X, Singer DB, Murdoch FE, Fritsch MK. The role of histone acetylation in regulating early gene expression patterns during early embryonic stem cell differentiation. J Biol Chem 2007; 282:6696 - 706; http://dx.doi.org/10.1074/jbc.M609519200; PMID: 17204470
- Yoo EJ, Chung JJ, Choe SS, Kim KH, Kim JB. Down-regulation of histone deacetylases stimulates adipocyte differentiation. J Biol Chem 2006; 281:6608 - 15; http://dx.doi.org/10.1074/jbc.M508982200; PMID: 16407282
- Frye M, Fisher AG, Watt FM. Epidermal stem cells are defined by global histone modifications that are altered by Myc-induced differentiation. PLoS One 2007; 2:e763; http://dx.doi.org/10.1371/journal.pone.0000763; PMID: 17712411
- Saunders N, Dicker A, Popa C, Jones S, Dahler A. Histone deacetylase inhibitors as potential anti-skin cancer agents. Cancer Res 1999; 59:399 - 404; PMID: 9927053
- Elder JT, Zhao X. Evidence for local control of gene expression in the epidermal differentiation complex. Exp Dermatol 2002; 11:406 - 12; http://dx.doi.org/10.1034/j.1600-0625.2002.110503.x; PMID: 12366693
- Wellen KE, Hatzivassiliou G, Sachdeva UM, Bui TV, Cross JR, Thompson CB. ATP-citrate lyase links cellular metabolism to histone acetylation. Science 2009; 324:1076 - 80; http://dx.doi.org/10.1126/science.1164097; PMID: 19461003
- Sen GL, Webster DE, Barragan DI, Chang HY, Khavari PA. Control of differentiation in a self-renewing mammalian tissue by the histone demethylase JMJD3. Genes Dev 2008; 22:1865 - 70; http://dx.doi.org/10.1101/gad.1673508; PMID: 18628393
- Kaelin WG Jr., McKnight SL. Influence of metabolism on epigenetics and disease. Cell 2013; 153:56 - 69; http://dx.doi.org/10.1016/j.cell.2013.03.004; PMID: 23540690
- Lu C, Thompson CB. Metabolic regulation of epigenetics. Cell Metab 2012; 16:9 - 17; http://dx.doi.org/10.1016/j.cmet.2012.06.001; PMID: 22768835