Abstract
Apical lumen formation is a key step during epithelial morphogenesis of tubular organs. Appropriate transport and targeting of apical proteins to the apical membrane initiation site (AMIS) plays a crucial role in establishing a solitary, central lumen. FIP5, a Rab11-interacting protein, is an important regulator that directs apical endosome trafficking along microtubules toward the AMIS during cytokinesis. However, it is unknown which molecular motor(s) transports FIP5-positive apical endosomes during lumen initiation, and how this process is regulated. In this study, we demonstrate that the interaction of FIP5 with the microtubule motor, Kinesin-2, is required for the movement of FIP5-endosomes and delivery of these endosomes from centrosomes to the cleavage furrow during apical lumen initiation. Loss of Kinesin-2 disrupts targeting of apical proteins to the AMIS and results in multiple lumen formation in MDCK cysts. Our data provide more details to the molecular mechanism of FIP5-dependent apical trafficking during apical lumen formation.
Introduction
Epithelium lines the external and internal surfaces of organs in animals. To protect the interior of the body from external insults, the epithelium adopts an asymmetric, or polarized organization. In polarized epithelial cells, the apical and basolateral domains define the asymmetry of the plasma membrane (PM) and contain different proteins and lipids.Citation1 While the basolateral PM interacts with neighboring cells and the extracellular matrix (ECM), the apical surface faces the outside world in the skin or the lumen in hollow organs such as the kidney and intestine.Citation2,Citation3 During development of these tubular, hollow organs, it is key to form a single apical lumen surrounded by a monolayer of polarized epithelial cells. This process involves apical membrane specification and determination of the apical lumen initiation site, which requires appropriate apical protein targeting.Citation1
Recently a model for apical lumen formation has been proposed, in which an apical membrane initiation site (AMIS) is formed at the center of forming cysts at early stages of lumenogenesis.Citation4 Endocytic vesicles carrying apical proteins are then transported to and fuse at the AMIS, which gives rise to a nascent apical lumen. Rab11 small GTPase is required for proper lumen formation in MDCK cells and has been implicated in different pathways of endocytic trafficking, including protein transport to the forming apical lumen. To fulfill these different roles, Rab11 associates with various effector proteins including actin motor proteins such as Myosin Vb, as well as various scaffolding and tethering proteins.Citation5 FIPs, the Family of Rab11-Interacting Proteins, are a group of scaffolding proteins that have been indicated in regulating endocytic membrane trafficking.Citation6,Citation7 Among the five members of the FIP family, FIP5 interacts with Rab11 in polarized epithelial cells and plays an important role in apical protein targeting.Citation8,Citation9 FIP5 is required for proper epithelial lumen formation and loss of FIP5 results in the development of multiple apical lumena in MDCK cysts, which is likely due to mis-targeting of apical proteins at early stages of lumen initiation.Citation9 What remains unclear is how FIP5-containing apical endocytic carriers are transported and targeted to the forming AMIS during apical lumen initiation.
Microtubule motors play important roles in rapid and long distance cargo transport. A major group of microtubule motors is the kinesin superfamily. While some kinesins can transport cargo toward the minus-end of microtubules, the majority of kinesins motors are plus-end directed. We recently showed that Kinesin-2 binds to FIP5 and regulate endocytic recycling in HeLa cells.Citation10 In this study, we investigate whether the FIP5/Kinesin-2 complex is required for apical endocytic carrier targeting and transport to the AMIS during apical lumen formation. We demonstrate that Kinesin-2 is a major microtubule motor mediating apical endosome traffic in polarized epithelial cells. Additionally, we show that Kinesin-2 is required for single apical lumen formation and that Kinesin-2 transports FIP5-endosomes along central spindle microtubules toward the cleavage furrow, where the AMIS is formed during early stages of apical lumen initiation.Citation11 Thus, we propose that the interaction of FIP5 and Kinesin-2 mediates directional traffic of apical endocytic carriers along microtubules to the cleavage furrow during cytokinesis, and that this transport is required for apical protein transport to the lumen initiation site.
Results
Characterization of FIP5 and Kinesin-2 binding
Our previous studies suggested that FIP5 binds to Kinesin-2.Citation10 However, the specificity of this interaction has not been tested. Since Kif5B (Kinesin-1 subunit) and Kif13A were also shown to be involved in endosomal transport, we therefore first investigated whether FIP5 can bind to Kif5 or Kif13A. To that end, we performed a series of glutathione bead pull-down assays with purified 6His-FIP5 and various Kinesin subunits tagged with GST. As shown in , FIP5 binds to Kinesin-2 subunits Kif3A and Kif3B, but not Kif13A or Kif5B. The interaction of FIP5 with another subunit of Kinesin-2, Kif17, was weaker than FIP5′s interaction with Kif3A or Kif3B, and could not be observed when the tail domain of Kif17 alone was used (). Thus, while we cannot fully rule out that FIP5 may also bind to Kif17, these results indicate that Kinesin-2 consisting of Kif3A/Kif3B subunits is likely a primary Kinesin motor that directly interacts with and transports FIP5-endosomes. We also mapped the domains that mediate the interaction between FIP5 and Kinesin-2. As shown in , Kif3B aa621–747 is the minimal motif required for the binding of FIP5 and Kinesin-2 (also see Fig. S1).
Figure 1. Characterization of FIP5 and Kinesin-2 binding. (A) GST pull-down assays show that FIP5 specifically binds to Kinesin-2 subunits Kif3A and Kif3B at the tail domain (GST-Kif3A-T and GST-Kif3B-T), while the bindings of FIP5 to tail domains of Kif17, Kif5B and Kif13A are non-comparable. (B) The summary of domain mapping analysis using GST pull-down assays. (C) Kinesin-2 and SNX18 compete for binding to FIP5. The graph shows quantification of FIP5 and GST-Kif3A-T binding. The data shown are means and standard deviations from three independent experiments. (D) Various concentrations of recombinant purified 6His-FIP5 were incubated with glutathione beads coated with either GST-Kif17-CT or GST-Kif3A-T. The graph shows quantification of 6His-FIP5 binding to GST-Kif3A-T or GST-Kif17-CT. (E) FIP5 was immunoprecipitated from MDCK cell lysates with anti-FIP5 antibody in the presence or absence of 20 μg of GST-Kif3A-T protein. The precipitate was then immunoblotted for FIP5 (top blot) or SNX18 (bottom blot). Note that in MDCK cells FIP5 exists in two different splice isoforms. Numbers at the bottom of the blot are quantifications of the amount of SNX18 co-precipitating with anti-FIP5 antibody. The data shown are means and standard deviations derived fro three independent experiments.
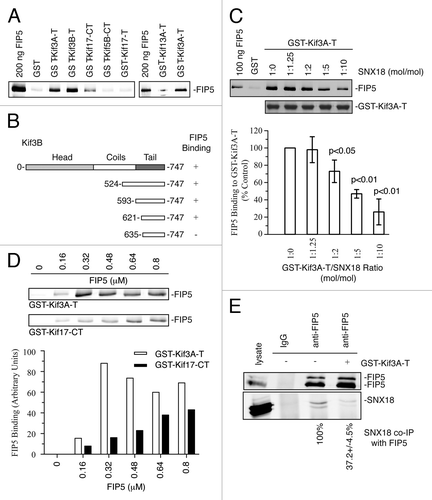
Previous studies identified SNX18 as a FIP5-binding protein that is required for the budding of FIP5-endosomes during apical lumen formation.Citation9 What remains unclear is whether SNX18 and Kinesin-2 can bind to FIP5 simultaneously. To answer this question, we tested GST-Kif3A binding to 6His-FIP5 in the presence or absence of purified recombinant SNX18 (). Increasing the concentration of SNX18 led to reduced binding of FIP5 to Kif3A, indicating that SNX18 and Kinesin-2 bind to overlapping motifs of FIP5 and likely compete for binding to FIP5. Consistently, the presence of the Kif3A-tail reduced co-immunoprecipitation of endogenous SNX18 with anti-FIP5 antibodies from MDCK cell lysates ().
Kinesin-2 mediates FIP5-endosome transport in MDCK cells
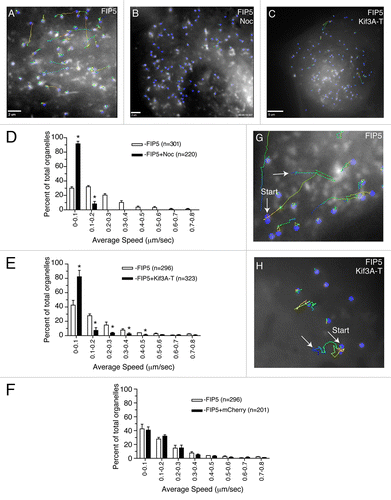
To determine whether Kinesin-2 transports FIP5-endosomes in MDCK cells, we transduced MDCK cells with an adenovirus expressing FIP5-GFP, and analyzed the mobility of FIP5-endosomes by time-lapse microscopy. As shown in , FIP5-endosomes were very dynamic, with the majority of the endosomes traveling at the speed of 0.1–0.5 μm/sec. This trafficking was dependent upon microtubules, since incubating cells with 10 μM nocodazole for 30 min significantly decreased the motility of FIP5-endosomes (). To determine whether Kinesin-2 is a motor that mediates FIP5-endosome trafficking, we performed time-lapse microscopy with MDCK cells co-expressing FIP5-GFP and mCherry-tagged Kif3A-Tail (mCherry-Kif3A-T), known to act as a dominant negative mutant.Citation12,Citation13 mCherry-Kif3A-T overexpression significantly reduced the mobility of FIP5-endosomes (). In contrast, overexpression of mCherry alone, did not have any effect on FIP5-endosome speed (). Furthermore, in mCherry-Kif3A-T co-expressing cells FIP5-endosome movement was predominately comprised of localized oscillations, in comparison to the linear trajectory seen in the cells co-expressing mCherry alone ().
Figure 2. FIP5-endosomes are transported by Kinesin-2. (A–H) MDCK cells were transduced with adenovirus expressing FIP5-GFP alone (A, B and G) or co-transfected with mCherry-Kif3A-T (C and H) or mCherry (F). Where indicated (B), cells were treated with 10 μM nocodazole for 30 min. Cells were plated on collagen-coated glass coverslips and the motility of the FIP5-GFP endosomes was analyzed using time-lapse microscopy. Panels (A, B, C, G and H) show the organelle trajectory over 35 s. Panels (D–F) show the quantification of the organelle speed. The data shown are the means from two independent experiments using three different cells. n is the number of organelles analyzed. Asterisks mark the bars that are different fro the control at P < 0.01. Scale bars: 2 μm (A and B), 5 μm (C).
Kinesin-2 transports FIP5-ednosomes along central spindle microtubules to the AMIS
We have recently shown that AMIS forms around the midbody during late telophase, and that cytokinesis-dependent AMIS formation is required for single apical lumen establishment.Citation11 Additionally, we and others have also shown that upon midbody-dependent AMIS formation, FIP5-endosomes are transported from centrosomes to the apical lumen formation site.Citation14 Since FIP5 binds to Kinesin-2, we hypothesized that FIP5-endosomes may be targeted to the midbody-associated AMIS by Kinesin-2-dependent transport along central spindle microtubules. To test this possibility, we first analyzed FIP5-GFP distribution in Matrigel-embedded cells treated with 10 μM of nocodazole for 45 min. It has been previously shown that nocodazole treatment causes depolymerization of dynamic microtubules, while having no effect on acetylated microtubules, such as the central spindle.Citation15,Citation16 Consistent with the previous studies using filter-grown MDCK cells,Citation8 nocodazole treatment in interphase cells caused dispersal of FIP5-containing apical endosomes, while maintaining their enrichment at the apical pole (). In contrast, nocodazole treatment had no effect on translocation of FIP5-endosomes to the cleavage furrow during late telophase (). Under these conditions nocodazole does not affect central spindle microtubules (), therefore, we propose that FIP5-endosomes are transported along the central spindle to the midbody/AMIS.
Figure 3. Kinesin-2 transports FIP5-endosomes along central spindle microtubules to the AMIS. (A–D) MDCK cells expressing FIP5-GFP were embedded into Matrigel and grown for 24 h. Cells were then incubated for 30 min in the presence (B–D) or absence (A) of 1 μM nocodazole, fixed and stained for either β-tubulin (D) or acetylated tubulin (C). Arrows point to either the lumen (A–B) or midbody (C–D). Scale bars: 5 μm. (E–F) MDCK cells expressing FIP5-GFP were embedded into Matrigel and plated on glass-bottom dishes. After incubation for 24 h, cells in early telophase (E–F) were chosen for time-lapse analysis. To depolymerize microtubules, 10 μM of nocodazole was then added and cells were imaged every five minutes for 90 min at 37 °C. Panels in (F) show selected images from time-lapse series. Arrow points to the midbody location. Scale bars: 5 μm. (G) MDCK cells transiently co-transfected with FIP5-GFP and mCherry-Kif3A-T were grown in 3D cultures for 24 h. Dividing cells were then analyzed by time-lapse microscopy. Asterisks mark FIP5 associated with centrosomes. Scale bars: 5 μm. (H–L) MDCK-shKif3A#1 cells were pre-incubated with or without doxycycline for 3 d and then grown in 3D cultures for 24 h in the presence (I and K) or absence (H and J) of doxycycline. Cells were then fixed and stained with anti-gp135 (H and I), anti-FIP5 (H and I) or anti-Rab11 (J and K) antibodies. Scale bars: 10 μm. Figure in (L) shows line-scan quantifications of Rab11 distributions (marked by lines in J and K).
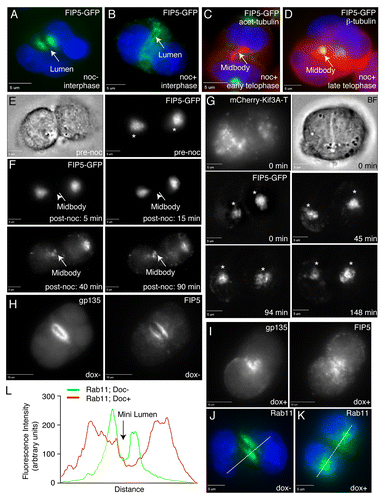
To determine whether Kinesin-2 is required for the movement of FIP5-endosomes from centrosomes to the midbody in MDCK cells, we analyzed FIP5-GFP dynamics in MDCK cells co-expressing FIP5-GFP and mCherry-Kif3A-T. As shown in , Kif3A-T dominant-negative mutant prevented FIP5 translocation, while having no effect on FIP5 accumulating around centrosomes during metaphase and anaphase.
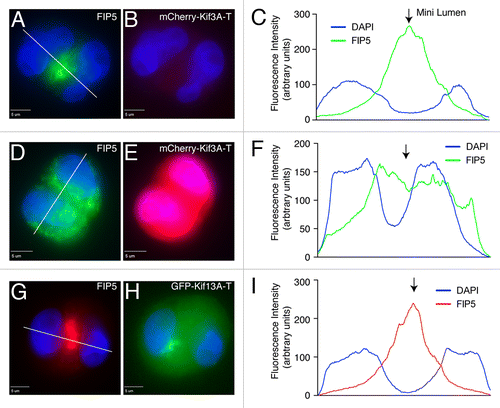
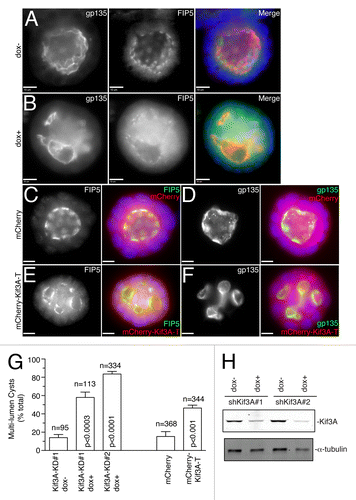
All the data shown above suggest that Kinesin-2 is required for apical lumen formation by mediating transport of apical cargo to the AMIS during cytokinesis. To further test this, we used an MDCK cell line stably expressing doxycycline-inducible Kif3A shRNA (MDCK-shKif3A#1),Citation17 since both Kif3A and Kif3B subunits are required to form a functional Kinesin-2 motor in epithelial cells. We co-stained MDCK-shKif3A cysts grown in the presence or absence of doxycycline with anti-gp135 (an apical PM marker), anti-FIP5 or anti-Rab11 antibodies 24 h after embedding cells in 3D cultures. We have previously shown that 24 h after embedding MDCK cells in Matrigel, the majority of cells are in the two-cell stage with well-formed mini-lumen.Citation9 Consistent with that, in the absence of doxycycline most cells formed clearly identifiable nascent lumens, with FIP5 and Rab11-endosomes accumulating at the apical pole of the cysts (). In contrast, Kif3A knockdown by the addition of doxycycline led to the dispersed distribution of FIP5 and Rab11 in the cytoplasm and a lack of nascent lumen formation (). To further confirm that Kinesin-2 is required for the transport of FIP5-endosomes to the site of lumen formation we analyzed Matrigel-embedded cells expressing either mCherry-Kif3A-T or GFP-Kif13A-T (). Consistent with our knock-down data, mCherry-Kif3A-T but not GFP-Kif13A-T blocked the accumulation or FIP5-endosomes at the lumen formation site ().
Figure 4. Kinesin-2 is required for FIP5-endosome targeting to the AMIS. Control MDCK cells (A–C) or MDCK cells transiently expressing either mCherry-Kif3A-T (D–F) or GFP-Kif13A-T (G–I) were grown in 3D cultures for 24 h. Cells were then fixed and stained with anti-FIP5 antibodies. Panels in (C, F and I) show quantification of FIP5 distribution (marked by lines in A, D and G). Scale bars: 5 μm.
Kinesin-2 is required for epithelial lumen formation
Since Kinesin-2 is required for apical protein transport during apical lumen initiation, it would also be expected that Kinesin-2 is required for the formation of single apical lumen in mature epithelial cysts. To test this hypothesis, MDCK-shKif3A cells were grown in 3D cultures in the presence or absence of doxycycline for 5 d to allow the formation and expansion of the apical lumen. Consistent with the involvement of Kinesin-2 in single apical lumen formation, Kif3A knock-down using two different shRNAs led to the formation of multi-luminal cysts, as shown by staining with anti-FIP5 and anti-gp135 antibodies (), while having no effect on overall distribution of microtubules and Trans-Golgi Network (TGN) (Fig. S2). To further confirm the requirement of Kinesin-2, we transfected MDCK cells with Kinesin-2 dominant-negative mutant mCherry-Kif3A-T. Confirming the role of Kinesin-2 in lumen formation, mCherry-Kif3A-T overexpression also caused a multi-luminal phenotype (), while overexpression of mCherry alone had no effect on epithelial cyst formation (). Taken together, these data demonstrate that Kinesin-2 is required for normal epithelial morphogenesis, likely by mediating the transport of FIP5-endosomes along the central spindle during initial stages of apical lumen initiation.
Figure 5. Kinesin-2 is required for apical lumen formation. (A–B) MDCK-shKif3A#1 cells were grown in 3D cultures for 5 d in the presence (B) or absence (A) of doxycycline. Cells were then fixed and stained with anti-gp135 and anti-FIP5 antibodies. Scale bars: 10 μm. (C–F) MDCK cells transiently transfected with either mCherry (C–D) or mCherry-Kif3A-T (E–F) were grown in 3D cultures for 5 d. Cells were then fixed and stained with anti-gp135 and anti-FIP5 antibodies. Scale bars: 10 μm. (G) Quantifications of the number of multi-luminal cysts in MDCK-shKif3A#1, MDCK-shKif3A#2, and MDCK cells transiently transfected with mCherry or mCherry-Kif3A-T. The data shown are means and standard deviations from three independent experiments. n is the total number of cysts counted. (H) MDCK-shKif3A#1 and MDCK-shKif3A#2 cells were grown in the presence or absence of doxycycline. Cells were then harvested and lysates were immunoblotted for the presence of Kif3A and α-tubulin.
Discussion
Apical lumen formation is a key step to epithelial morphogenesis.Citation4,Citation9,Citation14,Citation18 The establishment and maintenance of a single apical lumen requires targeted apical membrane trafficking, in which the Rab11 small GTPase is known to be an important regulator.Citation4,Citation19,Citation20 Although studies have shown that the interaction of Rab11 with its effector protein FIP5 is required for apical endocytic recycling and lumen formation, limited data are available for fully understanding how the Rab11/FIP5 complex regulates endocytic transport.Citation8,Citation9 In this study, we focused on characterizing the interaction of FIP5 with Kinesin-2, and investigating how this interaction contributes to apical lumen initiation. We show that Kinesin-2 binding to FIP5 mediates the delivery of FIP5-endosomes along central spindle microtubules to the AMIS during cytokinesis and is required for apical lumen formation.
While the Rab11/FIP5 complex was shown to mediate the formation of apical endocytic carriers, the machinery that regulates the transport and targeting of these carriers to the AMIS remains unclear.Citation9 Previous work has shown that FIP5 binds to the Kinesin-2 molecular motor in non-polarized HeLa cells.Citation10 Thus, we hypothesized that FIP5-endosome transport to the AMIS is microtubule-dependent and is mediated by Kinesin-2. In support of this hypothesis, our data demonstrate that FIP5 binds to Kinesin-2, but not Kinesin-1 or Kif13A, at the tail domain. The specificity in cargo/motor combination is one of the crucial factors that ensure the fidelity of membrane trafficking. While our results demonstrate that Kif3A/Kif3B heterodimer of Kinesin-2 is a primary FIP5-endosome motor, we were able to detect the binding weaker binding of FIP5 to the Kif17, suggesting that Kif17 motor may also play a role in apical lumen formation. Further investigations will be needed to characterize the interaction between FIP5 and Kif17 and elucidate the role of their binding in lumen formation.
Previous work has shown that FIP5 also binds sorting nexin 18 (SNX18). SNX18 and FIP5 interaction is required for apical endocytic carrier formation and lumen formation.Citation9 What remained unclear is whether Kinesin-2 and SNX18 can simultaneously bind to FIP5 and function as part of a large protein complex. Interestingly, our data show that Kinesin-2 and SNX18 compete for binding to FIP5, suggesting that FIP5 forms mutually exclusive complexes with SNX18 and Kinesin-2. This observation is consistent with the previous work demonstrating that FIP5-T276 phosphorylation by GSK-3 inhibits FIP5 binding to SNX18, while having no effect on FIP5 and Kinesin-2 interaction.Citation11 Taken together, these data suggest that FIP5 regulates apical protein trafficking by binding to various partners in a step-wise manner ().
Figure 6. Models for Kinesin-2-directed protein transport during apical lumen initiation. (A) Model for apical endosomal transport along central spindle microtubules at late stages of mitosis during apical lumen formation. (B) Proposed mechanism for Kinesin-2/FIP5-mediated FIP5-endosome trafficking during cytokinesis.
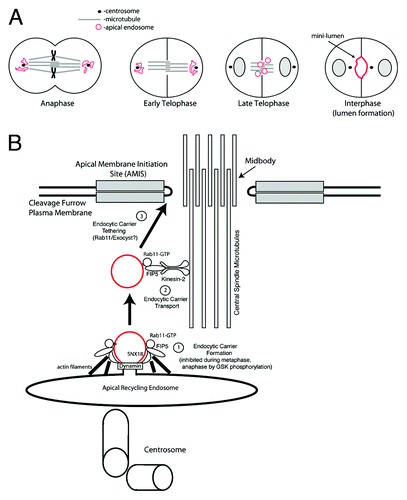
Previous studies have suggested that cytokinesis plays a role in the establishment of an apical lumen.Citation14 The AMIS forms during late telophase around the midbody, thus determining the apical lumen initiation site.Citation11 During anaphase and early telophase apical endosomes accumulate around centrosomes ().Citation11 During late telophase, after formation of the midbody-associated AMIS, apical endosomes translocate from centrosomes to the cleavage furrow, where they fuse with AMIS to establish the apical lumen ().Citation11 Interestingly, during the ingression of the cytokinetic cleavage furrow, central spindle microtubules overlap and are densely packed within the cleavage furrow with their plus-ends embedded in the midbody. This makes the central spindle microtubules perfectly suited to mediate the Kinesin-2-dependent transport toward the midbody and could provide spatial direction for apical protein transport to the AMIS. Since central spindle microtubules are resistant to nocodazole treatment, we used nocodazole to demonstrate that during late telophase FIP5-endosomes are primarily transported along central spindle microtubules. We also show that depletion of functional Kinesin-2 via the expression of Kif3A shRNAs or a Kif3A dominant-negative mutant reduces the transport of FIP5-endosomes to the AMIS along central spindle microtubules. Finally, we demonstrate that loss of Kinesin-2 leads to the formation of multi-luminal cysts, an indication of defects in epithelial apical lumen.
We therefore propose that during epithelial lumen initiation, Kinesin-2 interacts with FIP5 and delivers FIP5-endosomes carrying apical proteins along central spindle microtubules to the midbody, where the AMIS is located (). First, FIP5-endocytic carrier formation is mediated by FIP5 and SNX18 complex (), the process that is inhibited during anaphase by FIP5-T276 phosphorylation.Citation9 The FIP5-endocytic carriers are then handed over to Kinesin-2 via the FIP5/Kinesin-2 interaction. Central spindle microtubules then provide a spatial cue for the Kinesin-2-dependent trafficking of FIP5-endosomes toward the midbody-associated AMIS (). Finally, FIP5-endosomes fuse with AMIS to initiate apical lumen ().
In addition to Kinesin-2, several other microtubule motors have been shown to associate with endosomes. Recently published work by Dr. G. Raposo and colleagues has demonstrated that Kif13A can interact with Rab11 and regulate recycling endosome morphogenesis.Citation21 Kif16B was proposed to be required for transcytosis in epithelial cells.Citation22 Finally, it was demonstrated that Kinesin-1 transport transferrin receptor-containing recycling organelles during cell division.Citation23 Thus, it is clear that recycling endosome function depends on the activity of multiple kinesins. It is likely that various kinesin molecules are required for the distinct recycling endosome trafficking pathways in different cell types and during different stages of cell cycle. It is also possible that some of these kinesins are functionally redundant. Additional studies will be needed to define the possible cross-talk between all these endocytic kinesins.
Although this study expands our understanding in the spatiotemporal regulation of FIP5-regulated apical trafficking, much remains to be elucidated. Since FIP5 interacts with SNX18 for vesicle formation and with Kinesin-2 for vesicle transport, it is tempting to speculate that a protein may interact with FIP5 specifically for vesicle docking/tethering. While several proteins, such as the Exocyst complex and Slp2a/4a, were implicated in apical endosome targeting,Citation24 it remains to be determined whether they also mediate endocytic targeting during cytokinesis. Recent studies have also shown that accumulation of PtdIns(4,5)P2 appears to play a key role in determining the location of the apical lumen.Citation25 Importantly, PtdIns(4,5)P2 was also shown to accumulate at the cleavage furrow during cytokinesis.Citation26 Thus, it is possible that PtdIns(4,5)P2 may also function in initiating the AMIS formation and apical lumen initiation. Finally, most studies in lumenogenesis have been done using in vitro 3D tissue culture system, thus, it remains to be demonstrated whether the same molecules also play a role in lumen establishment and expansion in vivo. Answering these questions will help us better understand molecular mechanisms of apical protein trafficking and lumen formation during epithelial tissue morphogenesis.
Materials and Methods
Antibodies
The following antibodies were used: rabbit polyclonal anti-Kif3A (Abcam, #ab11259), mouse monoclonal anti-gp135 (a gift from C.Yeaman at University of Iowa, Iowa city, IA, and G.Ojakian at State University of New York Downstate Medical Center, Brooklyn, NY), mouse monoclonal anti-α-tubulin (Sigma, #T6074), Alexa Fluor® 488 protein A conjugate (Molecular Probes, P11047), Alexa Fluor® 594 protein A conjugate (Molecular Probes, P11051), cell-permeant Hoechst DNA stain (Invitrogen), goat anti–mouse AffiniPure F(ab’)2 fragments (Jackson ImmunoResearch Laboratories, Inc.), IRDye 800CW donkey anti-rabbit IgG (Li-Cor, 926–32213), and IRDye 680 donkey anti-mouse IgG (Li-Cor, 926–32222). Rabbit polyclonal anti-SNX18 antibody was prepared as described previouslyCitation8 using recombinant purified SNX18. The antibodies were affinity purified using recombinant SNX18 or cingulin conjugated to Affigel (Bio-Rad Laboratories) and eluted with 0.1 M glycine buffer, pH 2.5. Rabbit anti-FIP5 and anti-FIP1 polyclonal antibodies have been described previously.Citation8,Citation27,Citation28
Expression constructs and protein purification
Rab11a, SNX18 and all truncation mutants of Kif3A, Kif3B, Kif17, Kif13A and Kif5B were expressed as GST fusion proteins using the pGEX-KG plasmid (GE Healthcare). GST fusion constructs were expressed and purified using the BL21-(FE3)-RIPL Escherichia coli (E. coli) strain as described previously.Citation29 In brief, E. coli were lysed using a French press and then incubated with glutathione agarose beads (Sigma-Aldrich). Beads were then washed and either eluted with 25 mM glutathione or cleaved with thrombin (GE Healthcare). Full-length human FIP5 was tagged with an N-terminal 6His tag followed by a tobacco etch virus cleavage site by subcloning into the baculovirus transfer vector, pVL1392. Co-transfection and amplification of recombinant baculovirus was conducted using BacPAK transfection reagents (BD) according to the manufacturer’s instructions.
Cell culture and immunofluorescence microscopy
MDCK cells were cultured in DMEM with 4.5 g/liter glucose, 5.84 g/liter L-glutamine, 10% FBS (Takara Bio Inc.), and supplemented with 100 IU/ml penicillin and 100 µg/ml streptomycin. 3D cultures of MDCK cells were done as previously described.Citation9,Citation30 Briefly, MDCK cells were mixed with growth factor reduced Matrigel (BD) and plated in 12 µl drops on 8-well slides and overlaid with 400 μl of media. The cells were incubated for the indicated periods of time to allow for the development of epithelial cysts to different stages. Epithelial cysts were then fixed and stained according to a modified previously published protocol.Citation9,Citation31 Briefly, cells were fixed with 3% paraformaldehyde for 20 min, permeabilized with PBS containing 0.5% Triton X-100 for 10 min, and quenched three times for 15 min each time with a glycine/PBS solution (130 mM NaCl, 7 mM Na2HPO4, 3.5 mM NaH2PO4, and 100 mM glycine). Cells were incubated in primary block (10% FBS, 130 mM NaCl, 7 mM Na2HPO4, 3.5 mM NaH2PO4, 7.7 mM NaN3, 0.1% BSA, 0.2% Triton X-100, and 0.05% Tween-20) for 4 h, followed by incubation in secondary block (primary block with 20 µg/ml goat anti–mouse F(ab’)2 fragments) for 1 h. After washing, cells were left overnight in primary block with primary antibody and Hoechst nuclear stain. Cells were then washed and incubated for 1 h with secondary antibody in primary block. Cells were washed, dried for 1 h, and mounted with VectaShield (Vector Labs).
Glutathione bead pull-down assays
All glutathione bead pull-down assays were done as described previously.Citation9,Citation27,Citation29 Briefly, glutathione beads (50 µl) were coated with 10 µg of GST fusion protein or GST alone and incubated with varying amounts of soluble protein in a final volume of 0.5 ml of reaction buffer (50 mM Tris buffer, pH 7.5, containing 300 mM NaCl, 5 mM BME, 0.1% Triton X-100, 0.1% bovine serum albumin, and 1 mM phenylmethylsulfonyl fluoride). Samples were incubated at room temperature (RT) for 1 h on a nutator with constant rotation. The samples were pelleted at 2,000 g for 3 min and washed three times with 1 ml of reaction buffer. Bound proteins were eluted with 1% SDS, analyzed by SDS-PAGE, and either stained with Coomassie blue or immunoblotted.
Nucleofection
Nucleofection assays were performed as previously described.Citation32 Briefly, MDCK-MIIR cells were passaged ~24 h prior to transfection. For each transfection, 2 × 106 cells were resuspended in 85 μL Nucleofection solution T and 19 μL supplement 1 containing 2 μg of construct, and electroporated at program T-23 of Amaxa® Nucleofector® II Device (Lonza). 500 μL of pre-warmed RPMI media were added to the cells immediately after electroporation.
Time-lapse microscopy
To analyze FIP5-endosome motility, cells expressing FIP5-GFP were plated on collagen-coated 22 mm glass coverslips and incubated for 24 h. Cells were then mounted on a heater (DH-35) (Warner Instruments, Hamden, CT) fitted with a TC-344B dual automatic temperature controller (Warner Instruments), and imaged at 37 °C using a 63X oil-immersion lens. For all time-lapse series, 50 consecutive images were taken at 200 ms exposure with time-lapse of 500 ms. Organelle speed was then analyzed using by using the Intelligent Imaging Innovations (Denver, CO) three-dimensional rendering and exploration software. Only organelles which were present in the first time-lapse image and which could be followed in at least 10 consecutive images were analyzed. Over 200 organelles from five randomly chosen cells were analyzed for every condition. In every case the location of the centroid of each organelle was analyzed after every time point and the distance traveled during entire time-lapse series calculated. The overall speed of each organelle was then calculated based on the total distance traveled and the duration of the time-lapse series. Statistical analysis was performed using GraphPad Prism (La Jolla, CA) using a student’s T-test with a two-tailed, 95% confidence interval to generate p values.
| Abbreviations: | ||
| AMIS | = | apical membrane initiation sites |
| ECM | = | extracellular matrix |
| FIP | = | Family of Rab11 Interacting Protein |
| gp135 | = | glycoprotein 135 |
| MDCK cells | = | Madin-darby canine kidney cells |
| PtdIns(4,5)P2 | = | phosphatidyl inositol 4,5-P2 |
| PM | = | plasma membrane |
| TGN | = | Trans-Golgi Network |
Additional material
Download Zip (196.5 KB)Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Irene Choi for critical reading of the manuscript. We are appreciative of Dr. Charles Yeaman (University of Iowa) for anti-gp135 antibody. We also thank Dr. Cedric Delevoye (Institut Curie) for providing GFP-Kif13A-tail construct. This work was supported by a grant from the NIH-NIDDK (DK064380 to RP).
Supplementary Material
Supplementary material may be downloaded here: www.landesbioscience.com/journals/cellularlogistics/article/28928/
References
- Apodaca G, Gallo LI, Bryant DM. Role of membrane traffic in the generation of epithelial cell asymmetry. Nat Cell Biol 2012; 14:1235 - 43; http://dx.doi.org/10.1038/ncb2635; PMID: 23196841
- Schlüter MA, Margolis B. Apical lumen formation in renal epithelia. J Am Soc Nephrol 2009; 20:1444 - 52; http://dx.doi.org/10.1681/ASN.2008090949; PMID: 19497970
- Belting HG, Affolter M. It takes guts to make a single lumen. Nat Cell Biol 2007; 9:880 - 1; http://dx.doi.org/10.1038/ncb0807-880; PMID: 17671457
- Bryant DM, Datta A, Rodríguez-Fraticelli AE, Peränen J, Martín-Belmonte F, Mostov KE. A molecular network for de novo generation of the apical surface and lumen. Nat Cell Biol 2010; 12:1035 - 45; http://dx.doi.org/10.1038/ncb2106; PMID: 20890297
- Kelly EE, Horgan CP, McCaffrey MW. Rab11 proteins in health and disease. Biochem Soc Trans 2012; 40:1360 - 7; http://dx.doi.org/10.1042/BST20120157; PMID: 23176481
- Hales CM, Griner R, Hobdy-Henderson KC, Dorn MC, Hardy D, Kumar R, Navarre J, Chan EK, Lapierre LA, Goldenring JR. Identification and characterization of a family of Rab11-interacting proteins. J Biol Chem 2001; 276:39067 - 75; http://dx.doi.org/10.1074/jbc.M104831200; PMID: 11495908
- Prekeris R, Davies JM, Scheller RH. Identification of a novel Rab11/25 binding domain present in Eferin and Rip proteins. J Biol Chem 2001; 276:38966 - 70; http://dx.doi.org/10.1074/jbc.M106133200; PMID: 11481332
- Prekeris R, Klumperman J, Scheller RH. A Rab11/Rip11 protein complex regulates apical membrane trafficking via recycling endosomes. Mol Cell 2000; 6:1437 - 48; http://dx.doi.org/10.1016/S1097-2765(00)00140-4; PMID: 11163216
- Willenborg C, Jing J, Wu C, Matern H, Schaack J, Burden J, Prekeris R. Interaction between FIP5 and SNX18 regulates epithelial lumen formation. J Cell Biol 2011; 195:71 - 86; http://dx.doi.org/10.1083/jcb.201011112; PMID: 21969467
- Schonteich E, Wilson GM, Burden J, Hopkins CR, Anderson K, Goldenring JR, Prekeris R. The Rip11/Rab11-FIP5 and kinesin II complex regulates endocytic protein recycling. J Cell Sci 2008; 121:3824 - 33; http://dx.doi.org/10.1242/jcs.032441; PMID: 18957512
- Li D, Mangan A, Cicchini L, Margolis B, Prekeris R. FIP5 phosphorylation during mitosis regulates apical trafficking and lumenogenesis. EMBO Rep 2014; 15:428 - 37; http://dx.doi.org/10.1002/embr.201338128; PMID: 24591568
- Insinna C, Humby M, Sedmak T, Wolfrum U, Besharse JC. Different roles for KIF17 and kinesin II in photoreceptor development and maintenance. Dev Dyn 2009; 238:2211 - 22; http://dx.doi.org/10.1002/dvdy.21956; PMID: 19384852
- Brown CL, Maier KC, Stauber T, Ginkel LM, Wordeman L, Vernos I, Schroer TA. Kinesin-2 is a motor for late endosomes and lysosomes. Traffic 2005; 6:1114 - 24; http://dx.doi.org/10.1111/j.1600-0854.2005.00347.x; PMID: 16262723
- Schlüter MA, Pfarr CS, Pieczynski J, Whiteman EL, Hurd TW, Fan S, Liu CJ, Margolis B. Trafficking of Crumbs3 during cytokinesis is crucial for lumen formation. Mol Biol Cell 2009; 20:4652 - 63; http://dx.doi.org/10.1091/mbc.E09-02-0137; PMID: 19776356
- Zink S, Grosse L, Freikamp A, Bänfer S, Müksch F, Jacob R. Tubulin detyrosination promotes monolayer formation and apical trafficking in epithelial cells. J Cell Sci 2012; 125:5998 - 6008; http://dx.doi.org/10.1242/jcs.109470; PMID: 23097046
- Quinones GB, Danowski BA, Devaraj A, Singh V, Ligon LA. The posttranslational modification of tubulin undergoes a switch from detyrosination to acetylation as epithelial cells become polarized. Mol Biol Cell 2011; 22:1045 - 57; http://dx.doi.org/10.1091/mbc.E10-06-0519; PMID: 21307336
- Boehlke C, Kotsis F, Patel V, Braeg S, Voelker H, Bredt S, Beyer T, Janusch H, Hamann C, Gödel M, et al. Primary cilia regulate mTORC1 activity and cell size through Lkb1. Nat Cell Biol 2010; 12:1115 - 22; http://dx.doi.org/10.1038/ncb2117; PMID: 20972424
- Jaffe AB, Kaji N, Durgan J, Hall A. Cdc42 controls spindle orientation to position the apical surface during epithelial morphogenesis. J Cell Biol 2008; 183:625 - 33; http://dx.doi.org/10.1083/jcb.200807121; PMID: 19001128
- Desclozeaux M, Venturato J, Wylie FG, Kay JG, Joseph SR, Le HT, Stow JL. Active Rab11 and functional recycling endosome are required for E-cadherin trafficking and lumen formation during epithelial morphogenesis. Am J Physiol Cell Physiol 2008; 295:C545 - 56; http://dx.doi.org/10.1152/ajpcell.00097.2008; PMID: 18579802
- Li BX, Satoh AK, Ready DF. Myosin V, Rab11, and dRip11 direct apical secretion and cellular morphogenesis in developing Drosophila photoreceptors. J Cell Biol 2007; 177:659 - 69; http://dx.doi.org/10.1083/jcb.200610157; PMID: 17517962
- Delevoye C, Miserey-Lenkei S, Montagnac G, Gilles-Marsens F, Paul-Gilloteaux P, Giordano F, Waharte F, Marks MS, Goud B, Raposo G. Recycling endosome tubule morphogenesis from sorting endosomes requires the kinesin motor KIF13A. Cell Rep 2014; 6:445 - 54; http://dx.doi.org/10.1016/j.celrep.2014.01.002; PMID: 24462287
- Perez Bay AE, Schreiner R, Mazzoni F, Carvajal-Gonzalez JM, Gravotta D, Perret E, Lehmann Mantaras G, Zhu YS, Rodriguez-Boulan EJ. The kinesin KIF16B mediates apical transcytosis of transferrin receptor in AP-1B-deficient epithelia. EMBO J 2013; 32:2125 - 39; http://dx.doi.org/10.1038/emboj.2013.130; PMID: 23749212
- Montagnac G, Sibarita JB, Loubéry S, Daviet L, Romao M, Raposo G, Chavrier P. ARF6 Interacts with JIP4 to control a motor switch mechanism regulating endosome traffic in cytokinesis. Curr Biol 2009; 19:184 - 95; http://dx.doi.org/10.1016/j.cub.2008.12.043; PMID: 19211056
- Gálvez-Santisteban M, Rodriguez-Fraticelli AE, Bryant DM, Vergarajauregui S, Yasuda T, Bañón-Rodríguez I, Bernascone I, Datta A, Spivak N, Young K, et al. Synaptotagmin-like proteins control the formation of a single apical membrane domain in epithelial cells. Nat Cell Biol 2012; 14:838 - 49; http://dx.doi.org/10.1038/ncb2541; PMID: 22820376
- Shewan A, Eastburn DJ, Mostov K. Phosphoinositides in cell architecture. Cold Spring Harb Perspect Biol 2011; 3:a004796; http://dx.doi.org/10.1101/cshperspect.a004796; PMID: 21576256
- Brill JA, Wong R, Wilde A. Phosphoinositide function in cytokinesis. Curr Biol 2011; 21:R930 - 4; http://dx.doi.org/10.1016/j.cub.2011.10.001; PMID: 22115464
- Peden AA, Schonteich E, Chun J, Junutula JR, Scheller RH, Prekeris R. The RCP-Rab11 complex regulates endocytic protein sorting. Mol Biol Cell 2004; 15:3530 - 41; http://dx.doi.org/10.1091/mbc.E03-12-0918; PMID: 15181150
- Wilson GM, Fielding AB, Simon GC, Yu X, Andrews PD, Hames RS, Frey AM, Peden AA, Gould GW, Prekeris R. The FIP3-Rab11 protein complex regulates recycling endosome targeting to the cleavage furrow during late cytokinesis. Mol Biol Cell 2005; 16:849 - 60; http://dx.doi.org/10.1091/mbc.E04-10-0927; PMID: 15601896
- Junutula JR, Schonteich E, Wilson GM, Peden AA, Scheller RH, Prekeris R. Molecular characterization of Rab11 interactions with members of the family of Rab11-interacting proteins. J Biol Chem 2004; 279:33430 - 7; http://dx.doi.org/10.1074/jbc.M404633200; PMID: 15173169
- Vieira OV, Gaus K, Verkade P, Fullekrug J, Vaz WL, Simons K. FAPP2, cilium formation, and compartmentalization of the apical membrane in polarized Madin-Darby canine kidney (MDCK) cells. Proc Natl Acad Sci U S A 2006; 103:18556 - 61; http://dx.doi.org/10.1073/pnas.0608291103; PMID: 17116893
- Debnath J, Muthuswamy SK, Brugge JS. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods 2003; 30:256 - 68; http://dx.doi.org/10.1016/S1046-2023(03)00032-X; PMID: 12798140
- Chen X, Macara IG. RNA interference techniques to study epithelial cell adhesion and polarity. Methods Enzymol 2006; 406:362 - 74; http://dx.doi.org/10.1016/S0076-6879(06)06026-5; PMID: 16472670
