Abstract
Isotretinoin (13-cis RA) is the most potent agent in the treatment of acne. Insights into its mechanism of action can lead to drug discovery of alternative compounds with comparable efficacy but improved safety. The goal of this study is to compare the temporal changes in gene expression in the skin of acne patients after 1 week and 8 weeks of treatment with isotretinoin. Microarray analysis was performed on skin biopsies taken from 8 acne patients prior to and at 8 weeks of treatment with isotretinoin. Results were compared with data obtained from 7 acne patients biopsied at one week of treatment in a prior study. Distinctly different patterns of gene expression were noted. At 8 weeks, genes encoding extracellular matrix proteins were up-regulated and numerous genes encoding lipid metabolizing enzymes were down-regulated. At one week, genes encoding differentiation markers, tumor suppressors and serine proteases were up-regulated. Only three genes were commonly down-regulated. The temporal changes in gene expression in patient skin noted with isotretinoin substantiate many previously reported effects of isotretinoin and other retinoids, suggesting a model wherein isotretinoin induces apoptosis leading to reduced sebaceous gland size, decreased expression of lipid metabolizing enzymes and increased matrix remodeling during acne resolution.
Introduction
Transcriptional profiling represents a powerful tool to examine changes in gene expression. When combined with advances in bioinformatics, transcriptional profiling can generate data that may target specific genes or pathways for future hypothesis-driven investigation. The goal of the present study is to gain insight into the potential pathways by which 13-cis RA exerts its clearing effect in acne by comparing the temporal changes in gene expression in the skin of acne patients during treatment with isotretinoin. Array analysis and immunohistochemistry was performed on skin biopsies that were taken from the backs of eight acne patients at baseline and after 8 weeks of isotretinoin therapy. These data were compared to results from the array analysis and histology of skin from patients treated for one week as has recently been reported.Citation1
Distinctly different patterns of gene expression were noted depending upon the duration of therapy. In particular, marked decreases in the expression of genes involved in lipid metabolism were found at 8 weeks, which is in agreement with the marked decrease in sebaceous gland size. This contrasts with the significant increases in genes encoding differentiation markers, tumor suppressors, serine proteases and serine protease inhibitors observed at 1 week of isotretinoin therapy.Citation1
By examining the changes in individual gene expression using bioinformatics to identify enriched protein domains, chromosomal locations and cellular pathways and to analyze gene promoters that are preferentially influenced by isotretinoin, we hope to gain a greater understanding of isotretinoin’s influence on the entire cell. Furthermore, a comparison between the changes in gene expression at 1 week and 8 weeks of treatment allows us to appreciate the complexity of changes that occur in the skin during the course of therapy.
Results
Patient selection.
Early studies indicate that 13-cis RA drastically decreased sebaceous gland size after 16 weeks of treatment;Citation2 therefore, we chose an 8-week time point to examine the change in skin histology and gene expression in patients receiving the treatment for their severe acne. A total of eight patients who were prescribed isotretinoin by their dermatologist for their severe acne were enrolled in the study after signing informed consent forms. Patients had 5-mm punch biopsies of uninvolved skin taken from their upper backs at baseline and 8 weeks of treatment. Due to scheduling issues, there was variation in the length of time at which the second biopsy was performed (9.12 ± 1.1 weeks). Information regarding patient age, gender, timing of biopsy and the dose of isotretinoin that patient was receiving at the time of the second biopsy are presented in .
Significant decreases in sebaceous gland size after 8 weeks of treatment.
Hematoxylin and eosin staining was performed on sections of patient skin from the upper back that was biopsied at baseline and at approximately 8 weeks of isotretinoin therapy (). At baseline, sebaceous glands were large and multi-lobulated. After 8 weeks of treatment, sebaceous glands were markedly reduced in size by 76% (4.17-fold) from baseline (p = 0.009) (). Glands lost their multi-lobular structure and sparse sebocytes were noted only in close proximity to hair follicles. This architecture and location closely mirrors the sebaceous glands in murine skin.Citation3 In comparison, hematoxylin and eosin stained sections from biopsies taken at one week of isotretinoin treatment were decreased in size by approximately 49%; although, this decrease was not statistically significant ().Citation1
Significant decreases in expression of genes that regulate lipid metabolism were noted after 8 weeks of isotretinoin therapy.
Gene array expression analysis was performed on patient skin biopsies at baseline and at 8 weeks of isotretinoin therapy in order to gain insight into putative pathways affected by 13-cis RA treatment. Using a false discovery rate (FDR) of 0.05 that corresponds to a 5% chance of false positive gene changes, 197 genes were significantly upregulated and 587 genes were significantly downregulated. Select genes that showed either an upregulation or downregulation of approximately two-fold or greater are listed in . For a complete listing of all significantly changed genes at 8 weeks, see Supplemental Data. The preponderance of genes that were downregulated at 8 weeks are involved in the metabolism of steroids, cholesterol and fatty acids, which is consistent with the known decreases in sebaceous gland lipid production induced by 13-cis RA. In comparison, the genes whose expression changed significantly after 1 week isotretinoin can be broadly categorized as tumor suppressors, protein processors and genes involved in the transfer or binding of ions, amino acids, lipids or retinoids.Citation1
From among the 42 significantly changed genes at 1 week and the 784 significantly changed genes at 8 weeks, only three genes were found in common. These three genes were downregulated two- to five-fold and include solute carrier family 26 member 3 (SLC26A3); phospholipase A2, group VII (PLA2G7); and phosphodiesterase 6A (PDE6A). SLC26A3 is a chloride/bicarbonate exchanger in the colon which helps mediate absorption of electrolytes and fluid.Citation4 Its function in the skin is unknown; although, its expression has been detected within eccrine sweat glands.Citation5 Another solute carrier family member (SLC24A5) has been identified as playing a prominent role in skin biology by regulating flux across the melanosome membrane in zebrafish and links to skin color in humans.Citation6 Phospholipase A2 family members are responsible for hydrolyzing cell membrane phospholipids to generate arachidonic acid.Citation7 Phosphodiesterase 6A (PDEA6) encodes the cyclic-GMP specific PDE6A alpha subunit, which is expressed in cells of the retinal rod outer segment.
QPCR verification of select genes from array analyses.
Sufficient RNA was available from patients 1, 2, 4 and 7 to verify changes in gene expression by QPCR for 3β-hydroxysteroid dehydrogenase (3βHSD1), HMG CoA reductase (HMGCR), phospholipase A2 group 7 (PLA2G7), insulin induced gene 1 (INSIG), carnitine acyltransferase (CRAT) and zinc finger binding protein 145 (ZBTB16) (). The direction and magnitude of the changes in expression for the selected genes are similar to those observed with gene array analysis.
Cluster analysis.
Using the computer software, dChip,Citation8 we performed hierarchical clustering of the entire set of genes whose expression was changed significantly at 1 week or 8 weeks of isotretinoin treatment. A two-way cluster analysis was done; the data was clustered by patient sample and also by genes exhibiting similar expression profiles. We found that the baseline samples clustered into groups that were separate and distinct from the treatment samples for both the 1 week and 8 week data sets (; see Suppl. Data for 8 week diagram). Clustering of the patient samples (columns) is indicated at the top of the diagram. Genes with higher correlation coefficients among the standardized gene expression values across samples are clustered together by rows. Therefore, genes in the same cluster share similar expression patterns. Of note, the lack of correlation between the 1-week and 8-week data sets illustrates that isotretinoin regulates the temporal expression of separate and distinct groups of genes.
Functional categorization of significantly changed genes.
Gene expression analysis revealed numerous genes significantly affected by 13-cis RA treatment. In order to determine if 13-cis RA preferentially influences a particular subset of genes at either time point, we categorized the genes using dChip computer software. Genes included on each array carry annotations that allow them to be grouped according to categories including “gene ontology,” “protein domains,” ”chromosomal location” and “pathway.” It is important to note that annotations in each of these categories are not available for all genes on the arrays. Each category contains predefined terms for classifying genes. After hierarchical clustering, dChip assessed the significance of all functional categories within the cluster tree. The 42 genes changed in the 1-week analysis and the 784 genes whose expression was significantly changed in the 8-week analysis were assessed for significant enrichment for any “gene ontology,” “protein domain,” “chromosomal location” and “pathway.” The p-value for this functional categorization is the probability of seeing x genes with a certain category occurring in a group of k genes at random, given n annotated genes on the array, of which m genes carry that specific annotation. Groups of genes mapping to a particular cluster with p < 0.001 were considered significant.
Within the 784 significantly changed genes at 8 weeks, we identified 98 gene ontology terms (data not shown), 21 protein domains, 0 chromosomal locations and 10 pathways that were significantly enriched (p < 0.001). Of the enriched protein domains (, italics), “anaphylotoxin/fibulin,” “fibronectin, type 1” and “collagen triple helix repeat” were significantly upregulated, while all other protein domains were significantly downregulated. In the analysis of pathways, genes involved in each of the 10 pathways () were downregulated. Interestingly, each of these 10 downregulated pathways is either directly involved in or linked to fatty acid and cholesterol metabolism, further supporting 13-cis RA’s role in sebum suppression. A subset of genes that mapped to these enriched pathways is indicated in italics in .
From the 42 significantly changed genes at 1 week, we identified five gene ontology terms, three protein domains and one chromosomal location that were enriched according to dChip (). The chromosomal location of 1q21 is the site of the epidermal differentiation complex and five genes on this locus are affected by isotretinoin: cellular retinoic acid binding protein 2 (CRABP2), S100A2, S100A7, S100A9 and involucrin (IVL). No “pathways” were enriched in the 1-week data set, most likely due to the small number of significantly changed genes (42) and the fact that only three of these genes carried ”pathway” annotations.
In comparing the functional classification of the significantly changed genes, no commonalities were found between 1-week and 8-week data with regard to “gene ontology,” “protein domain,” “chromosomal location” or “pathway.”
Promoter analysis of genes.
Retinoids are known to regulate gene transcription via binding nuclear retinoid receptors. Although, 13-cis RA is not known to interact with these receptors; it is known to isomerize to both all-trans retinoic acid (ATRA) and 9-cis retinoic acid (9-cis RA), which do bind these receptors.Citation9,Citation10,Citation11 We analyzed the promoter regions of all the genes whose expression changed significantly at 1 week or 8 weeks for retinoid receptor consensus sequences using the Transcriptional Element Search System (TESS) database.Citation12 The significantly changed genes containing retinoic acid receptor response elements (RAREs) or rexinoid receptor response elements (RXREs) in their promoters are indicated in and . Some genes did not have promoter sequences available from the Cold Spring Harbor Laboratory database and are indicated with N/A in each table. Of the genes significantly changed at 8 weeks (), 26 of 44 (59%) contained RAR or RXR consensus sequences. Of the 117 genes that mapped to the 10 enriched pathways in the 8-week analysis, 75% of these genes contained RAR or RXR response elements in their promoters (). In comparison, in the 1-week analysis, 21 of 42 (50%) of the significantly changed genes contained RAR or RXR consensus sequences (). Interestingly, all genes located on chromosome 1q21 whose expression changed contain consensus sequences for retinoic acid receptors.
The relative proportion of genes containing RAR or RXR consensus sequences is similar between the 1-week and 8-week data sets, 50% and 59%, respectively.
Discussion
The biological effects of 13-cis RA are complex and the pathways by which sebum is decreased and acne is improved have yet to be fully elucidated. Early studies in the 1980s demonstrated that 13-cis RA markedly diminishes the size and secretion of sebaceous glands after 16 weeks of isotretinoin treatment.Citation2 Our study demonstrates that 13-cis RA markedly decreases sebaceous gland size by 8 weeks of treatment, and a trend toward this reduction is apparent at 1 week. This is consistent with the observations that sebum secretion can be markedly reduced by 13-cis RA as early as 2 weeks.Citation13,Citation14 However, the sebaceous gland architecture returns to pre-treatment levels as early as 2 months after cessation of therapy and the rate of return is faster with the lower doses of isotretinoin.Citation15 Despite this observation, for unknown reasons, isotretinoin induces permanent remission of acne in a majority of cases. No study to date has examined the global changes in gene expression that accompany these histological changes in the skin of patients treated with 13-cis RA for an extended period of time (8 weeks). Understanding the influence of isotretinoin on specific genes and cellular pathways over time provides insight into its mechanism of action within the skin and sebaceous gland.
Using array analysis, we were able to demonstrate distinctly different patterns of gene expression at 1 week and 8 weeks of treatment in patients receiving isotretinoin for severe acne. Hundreds of genes were significantly changed after 8 weeks of isotretinoin therapy. The preponderance of genes that were downregulated involve lipid and sterol metabolism and are characteristically expressed, although not exclusively, within differentiated sebaceous glands.Citation16 These changes at 8 weeks are consistent with the noted reduction in sebaceous gland size and are likely due to the reduction in sebaceous gland volume. The majority of genes that were upregulated at 8 weeks encode structural proteins of the extracellular matrix such as collagens, fibulin and fibronectin. Increased expression of these genes is consistent with the known effects of retinoids in rebuilding extracellular matrix as reported in studies of naturally-aged and photoaged skin.Citation17,Citation18,Citation19
To understand the early actions of 13-cis RA on the sebaceous gland, patients were sampled at one week of treatment as previously reported.Citation1 The 1 week time point in that study was chosen based on reports from patients who noted that retinoid related side-effects (dry eyes, chapped lips) began to appear within the first week of treatment. In contrast to the 8 week data, the preponderance of genes whose expression changed significantly at one week were upregulated. These genes encode proteins that can be broadly categorized as tumor suppressors, protein processors and proteins involved in transfer or binding of ions, amino acids, lipids or retinoids. Increased expression of lipocalin 2 (LCN2), which encodes the neutrophil gelatinase associated lipocalin (NGAL), induces apoptosis, both in the sebaceous glands in patient skin and in cultured SEB-1 sebocytes.Citation1
The distinction between the patterns of gene expression induced by 13-cis RA at 1 week and 8 weeks is substantiated by the finding that no common genes were upregulated and only 3 genes (solute carrier family 26, member 3, phosphodiesterase 6A and phospholipase A2) were downregulated at both time points. One of these genes, phosphodiesterase 6A (PDEA6) encodes the cyclic-GMP specific PDE6A alpha subunit, which is expressed in cells of the retinal rod outer segment. Mutations in PDE6A have been identified as one cause of autosomal recessive retinitis pigmentosa, which is associated with night blindness.Citation20 Although night blindness is a known potential side effect of isotretinoin treatment, to date it has not been linked with changes in expression of PDE6A in rod cells. Phospholipase A2 (PLA2G7) is also downregulated by isotretinoin. PLA2 family members are responsible for hydrolyzing cell membrane phospholipids to generate arachidonic acid (AA), a known inflammatory mediator, and lysophospholipids.Citation7 Retinoids, including 13-cis RA, have been shown to inhibit PLA2 activity and prevent AA release in human synovial fluid.Citation21 This report, together with our array data, supports the hypothesis that the anti-inflammatory action of 13-cis RA may be due to inhibition of PLA2 expression or activity.
Approximately half of the genes whose expression was significantly changed at either 1 week or 8 weeks contain retinoic acid receptor response elements within their promoters. This suggests possible direct gene regulation through the classical nuclear retinoid receptor pathway. Although 13-cis RA itself doesn’t activate RAR or RXR, it is mostly isomerized to ATRA which can directly activate retinoic acid receptors.Citation9,Citation22 However, not all genes whose expression was changed by 13-cis RA contain RAREs, indicating that indirect regulation or regulation through alternative transcription factors is also possible. For example, retinoic acid-induced expression of reelin, which encodes an extracellular matrix serine protease, requires activation of an SP-1 site.Citation23
In depth analysis of each change in gene expression is a timeconsuming endeavor and provides limited information regarding the overall biological response to isotretinoin. To determine if particular gene categories, protein domains, pathways or chromosome locations are preferentially influenced by 13-cis RA, both microarray data sets were analyzed by dChip computer software. After 8 weeks isotretinoin, 3 of the 21 significantly enriched protein domains were upregulated: “anaphylotoxin/fibulin,” “fibronectin, type 1” and “collagen triple helix repeat.” This corresponds with the increased expression of individual genes that encode structural proteins of the extracellular matrix such as collagens, fibulin and fibronectin. Changes in these protein domains support the known fact that retinoids stimulate extracellular matrix deposition in wound healing and in aging skin.
Among the genes whose expression was significantly increased at 1-week, there was an increased representation of genes within chromosomal location 1q21. CHR1q21 is the location of the epidermal differentiation complex (EDC) whose genes encode proteins that are critical to terminal differentiation of the epidermis, such as, cornified envelope proteins (IVL and loricrin); CRABP2; S100 proteins; and filaggrin. Increased CRABP2 levels support a prior study demonstrating increased CRABP2 levels in patients’ sebaceous glands following 3–16 weeks of isotretinoin therapy as determined by in situ hybridization.Citation24 S100 proteins modulate cellular differentiation, energy metabolism, cytoskeletal membrane interactions, antimicrobial activities and cell cycle progression. Interestingly, the specific S100 proteins whose genes are upregulated in our array analysis are induced by oxidative or inflammatory stress. S100A7 (psoriasin) has antimicrobial effects against P. acnes in addition to functioning as a chemoattractant agent for immune cells.Citation25,Citation26 One can speculate that initial upregulation of S100 proteins by 13-cis RA could be responsible for the “acne-flare” response observed in some patients receiving oral or topical retinoids for the treatment of their acne. Our gene expression analysis of patient skin provides in vivo confirmation of the increase in S100 protein noted in cultures of normal human epidermal keratinocytes treated with 13-cis RA, 9-cis RA, ATRA and 4-oxo-13-cis RA.Citation27
Interestingly, in the 1-week array analysis of patient skin each of the significantly changed genes on chromosome 1q21 contained consensus sequences for retinoic acid receptors. Retinoids are crucial to normal epidermal development and differentiationCitation28 in addition to embryonic development through their effects on Hox gene expression.Citation29 Ichthyosis, atopic dermatitis and predisposition to psoriasis have all been linked to genetic variations within the EDC.Citation30 The mechanisms by which retinoids regulate genes within the EDC are of interest. The clustering of numerous retinoic-acid responsive genes on 1q21 raises the question as to whether each of these genes is individually transcribed following binding of retinoic acid to its promoter elements or whether retinoic acid affects epigenetic regulation of the EDC through a locus control region, similar to regulation of the β-globin gene locus on chromosome 11p15.5, or by other epigenetic mechanisms.Citation31
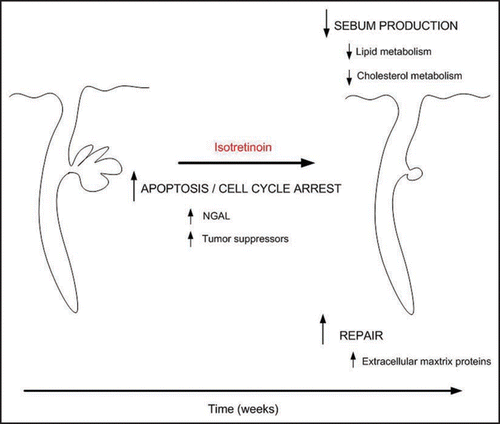
In summary, the temporal changes in gene expression in patient skin following treatment with isotretinoin substantiate many of the clinical, histological and biochemical effects of isotretinoin and other retinoids noted in previous studies. These data combined with recent findings regarding the effects of isotretinoin on apoptosis, cell cycle arrest and induction of NGAL expression suggest a putative model wherein isotretinoin exerts a rapid effect in inducing apoptosis and cell cycle arrest within the sebaceous gland that is, in part, mediated by expression of NGAL. This leads to a decrease in sebaceous gland size beginning at one week of treatment and continuing through 8 weeks of treatment, and perhaps beyond with a subsequent decrease in the expression of genes involved in lipid production. We hypothesize that after the initial induction of apoptosis and cell cycle arrest within the sebaceous gland, the skin adopts a wound healing-like pattern of gene expression and subsequently undergoes substantial repair and remodeling as evidenced by increased expression of extracellular matrix proteins (). What remains to be determined is the mechanism by which isotretinoin induces a long-term remission or permanent resolution of acne despite the return of the sebaceous gland function several months following completion of therapy.
Materials and Methods
Patient selection and tissue biopsies.
All protocols were approved by the Institutional Review Board of The Pennsylvania State University College of Medicine and were conducted according to the principles outlined in the Declaration of Helsinki. All subjects gave informed consent. The study included both males and females, ages 14 to 40 years, who were scheduled by their dermatologist to receive treatment with 13-cis RA (isotretinoin, brand not noted) for severe acne. All aspects of the patients’ treatment with 13-cis RA apart from the skin biopsies were standard of care and were not part of this research. Exclusion criteria included patients on medications that could alter sebum excretion such as hormonal therapy (including oral contraceptives) or patients with underlying medical conditions requiring treatment with systemic medications that might interfere with the gene array analysis. Patients had a 5-mm punch biopsy of skin taken from their upper back before treatment and after approximately 8 weeks of daily treatment with oral isotretinoin. (See for treatment details). Biopsies were placed on ice and immediately transferred to the laboratory where they were trimmed of fat and a small section of each biopsy was taken and paraffin-embedded for histology and immunohistochemistry. The remaining portion of the biopsy was flash frozen in liquid nitrogen and used for total RNA isolation.
Image analysis of sebaceous gland size.
Following hematoxylin and eosin staining of sections from each of the biopsies, image analysis of sebaceous gland size was performed. At least two baseline and two “after treatment” sections were analyzed from each subject. Images were captured using a Spot digital camera (Diagnostic Instruments, Inc.,) and measurements were obtained with Image Pro Plus Imaging Software Version 3.0 after spatial calibration with a micrometer slide under 10X magnification. All areas of sebaceous glands were circled using a free-hand measuring tool and the total area of the sebaceous gland was calculated in each section from baseline and at 8 weeks of treatment. Paired t-tests (α = 0.05) were performed to determine significant differences in sebaceous gland size before and after treatment.
Gene expression microarray analysis.
Total RNA was isolated from skin biopsies and DNase treated using the RNeasy Fibrous Tissue Kit (Qiagen Inc., Valencia, CA). Approximately 2 µg of total RNA from each sample was used to generate double stranded cDNA using a T7-oligo (dT) primer. Biotinylated cRNA, produced through in vitro transcription, was fragmented and hybridized to Affymetrix human U133A 2.0 microarrays. The arrays were processed on a GeneChip Fluidics Station 450 and scanned on an Affymetrix GeneChip Scanner (Santa Clara, CA). Expression signals were normalized as previously described.Citation32,Citation33,Citation34 Significant gene expression alterations were identified using Significance Analysis of Microarrays (SAM) computer software.Citation35 SAM controls the false positives resulting from multiple comparisons through controlling the false discovery rate (FDR).Citation36 FDR is defined as the proportion of false positive genes among all genes that are considered significant. The data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus (GEO, www.ncbi.nlm.nih.gov/geo/) and are accessible through GEO Series accession number GSE11792.
Quantitative real-time polymerase chain reaction (QPCR).
Quantitative real-time PCR experiments confirmed the direction and magnitude of changes in the expression of select genes from the microarray. Applied Biosystems’ Assays-on-Demand Taqman Universal PCR Master Mix, primer probe sets, and ABI’s 7900HT Fast Real-Time PCR System with 384-well plate block module were used according to manufacturer’s instructions (Applied Biosystems, Foster City, CA). Integrity of isolated RNA was verified by agarose gel electrophoresis. cDNA was generated from 1 µg of total RNA, primed with oligo-dT, using the Superscript First-Strand Synthesis System for reverse transcription- PCR (Invitrogen, Carlsbad, CA). Diluted cDNA samples were run for the reference gene TATA binding protein (TBP) and genes of interest: 3β-hydroxysteroid dehydrogenase (3βHSD1), HMG CoA reductase (HMGCR), phospholipase A2 group 7 (PLAG7), insulin induced gene 1 (INSIG), carnitine acyltransferase (CRAT) and zinc finger binding protein 145 (ZBTB16). Assay controls included samples omitting reverse transcriptase enzyme as well as samples without cDNA. Data was analyzed using the Relative Expression Software Tool (REST-XL version 1) software programCitation37 with efficiency correction and a p-value <0.05 was considered significant.
Cluster analysis.
Hierarchical clustering of patient samples and of significantly changed genes was performed using the normalized array data imported into dChip software version 1.3. The information files for the Affymetrix human genome HG-U133A 2.0 array was obtained from www.dChip.org. Each row represents a single gene and each column represents a patient sample. (B = baseline and A = after treatment). The color reflects the level of expression when compared to the mean level of expression for the entire biopsy set. Red indicates expression higher than the mean and blue indicates lower expression than the mean.
Database analysis of gene promoters.
The first 1,000 basepairs (promoter regions included) of the top 136 genes with the greatest statistically significant fold change at 8 weeks and all genes changed significantly at 1-week were examined for retinoic acid response elements (RAREs and RXREs). The base pair sequences of each gene were obtained from the Cold Spring Harbor Laboratories Promoter Database (ftp://cshl.edu/pub/science/mzhanglab/PromoterSet). These sequences were scanned for RAREs and RXREs using the Transcription Element Search System (TESS)Citation11 in conjunction with predefined consensus sequences within the TRANSFAC database.Citation38
Comparisons of gene expression arrays.
The 1-week and 8-week array data were compared to identify common genes whose expression was influenced by 13-cis RA. The data from the functional categorization of significantly changed genes was similarly compared to identify common gene ontologies, protein domains, chromosomal locations or pathways that are affected by isotretinoin treatment. A comparison was also made between the percentage of significantly changed genes containing RAR or RXR consensus sequences.
Abbreviations
| 13-cis RA | = | 13-cis retinoic acid |
| ATRA | = | all-trans retinoic acid |
| 9-cis RA | = | 9-cis retinoic acid |
| QPCR | = | quantitative real-time PCR |
Figures and Tables
Figure 1 Sebaceous gland size is significantly reduced by isotretinoin at 8 weeks of treatment compared to one week. Hematoxylin and eosin sections of skin biopsies were taken from patients before and at approximately 8 weeks or 1 week of isotretinoin treatment. (A) A total of 16 sections at baseline and 12 sections at 8 weeks of treatment were taken from 8 patients and analyzed using image analysis software. The mean area (±SEM) of sebaceous glands in the baseline samples was 0.17 ± 0.04 mm2 compared to 0.04 ± 0.07 mm2 in the 8-week samples; a four-fold reduction in size (p = 0.009). (B) A total of 17 sections at baseline and 19 sections at 1 week of treatment were taken from six patients and analyzed using image analysis software. In contrast to 8 weeks of isotretinoin treatment, the mean area (±SEM) of sebaceous glands in the 1-week baseline samples was 0.23 ± 0.09 mm2 compared to 0.12 ± 0.02 mm2 after 1 week of treatment, which was not significant using a paired t-test (p = 0.16). Representative images from three patients at each time point are shown at original magnification of 100x. Magnification bars = 250 µm. *Initial publication: Journal of Investigative Dermatology advance online publication, 6 November 2008; doi:10.1038/jid.2008.338.
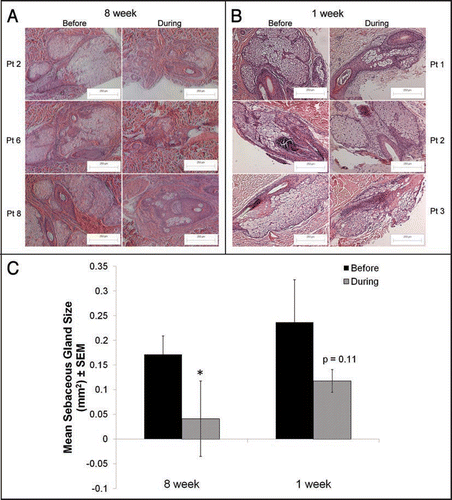
Figure 2 QPCR verifies the magnitude and direction of gene array changes. In order to verify the direction and magnitude of the changes in gene expression induced by isotretinoin in the gene array expression analysis, QPCR was performed using primers to select genes whose expression was significantly changed by 13-cis RA in the array analysis. Data represent the mean ± SEM of the fold-change in gene expression as determined by QPCR in four subjects compared to array analysis performed in eight subjects: 3β-hydroxysteroid dehydrogenase (3βHSD1), HMG CoA reductase (HMGCR), phospholipase A2 group 7(PLAG7), insulin induced gene 1 (INSIG), carnitine acyltransferase (CRAT) and zinc finger binding protein 145 (ZBTB16). Significant changes in gene expression with the microarray were identified using Significance Analysis of Microarrays (SAM) computer software and QPCR results were analyzed by the REST-XL software program; *p < 0.05 was considered significant for both analyses.
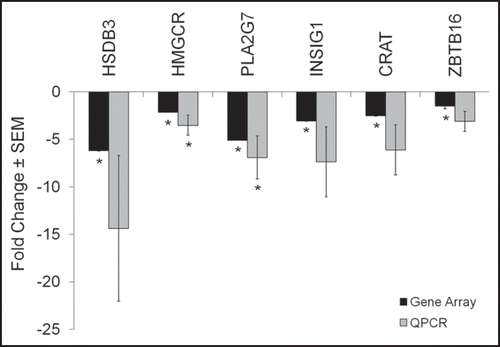
Figure 3 One week Clustering Diagram. Hierarchical clustering was used to compute a dendogram that assembled all samples and genes into a single tree. Normalized gene array expression data from each patient’s baseline and treatment biopsy was imported into dChip software version 1.3. Each row represents a single gene and each column represents a patient sample. B = baseline and A = after treatment. The color reflects the level of expression when compared to the mean level of expression for the entire 1 week biopsy set.
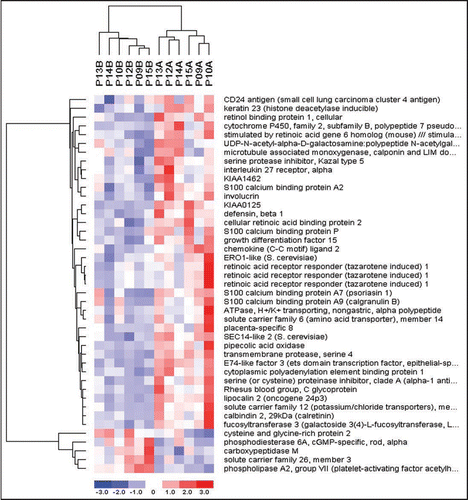
Table 1 8-week patient demographics
Table 2 Select genes changed significantly by isotretinoin in patient skin at 8 weeks
Table 3 Protein domains and pathways enriched within the genes significantly changed at 8 weeks
Table 4 Pathways enriched within the genes significantly changed at 8 weeks
Table 5 Functional categorization of significantly changed genes at 1 week isotretinoin
Table 6 Promoter analysis of genes significantly changed at 1 week of isotretinoin therapy
Additional material
Download Zip (707.3 KB)Acknowledgements
The authors wish to acknowledge Amy Longenecker and Silvia Gosik for coordination of patient procedures. This study was supported by NIH NIAMS R01 AR047820 to D.M.T. and NIH General Clinical Research Center Grants M01RR010732 and C06RR016499 to the Pennsylvania State University College of Medicine and by the Jake Gittlen Cancer Research Foundation and the Department of Dermatology at the Pennsylvania State University College of Medicine.
References
- Nelson A, Gililland K, Zhao W, Zaeglein A, Lui W, Thiboutot D. Neutrophil gelatinase associated lipocalin mediates 13-cis retinoic acid-induced apoptosis of human sebaceous gland cells. J Clin Invest 2008; 118:1468 - 1478
- Goldstein JA, Comite H, Mescon H, Pochi PE. Isotretinoin in the treatment of Acne. Arch Dermatol 1982; 118:555 - 558
- Picton W, Clark C. The effect of various treatments on the size of sebaceous glands of hairless mice and hairless hamsters. Br J Dermatol 1977; 96:277 - 282
- Hoglund P, Haila S, Socha J, Tomaszewski L, Saarialho-Kere U, Karjalainen-Lindsberg M-L, et al. Mutations of the Downregulated in adenoma (DRA) gene cause congenital chloride diarrhoea. Nat Genet 1996; 14:316 - 319
- Makela S, Kere J, Holmberg C, Hoglund P. SLC26A3 mutations in congenital chloride diarrhea. Hum Mutat 2002; 20:425 - 438
- Lamason RL, Mohideen MA, Mest JR, Wong AC, Norton HL, Aros MC, et al. SLC24A5, a putative cation exchanger, affects pigmentation in zebrafish and humans. Science 2005; 310:1782 - 1786
- Yedgar S, Cohen Y, Shoseyov D. Control of phospholipase A2 activities for the treatment of inflammatory conditions. Biochim Biophys Acta 2006; 1761:1373 - 1382
- Li C, Hung Wong W. Model-based analysis of oligonucleotide arrays: model validation, design issues and standard error application. Genome Biol 2001; 2:32
- Allenby G, Bocquel MT, Saunders M, Kazmer S, Speck J, Rosenberger M, et al. Retinoic acid receptors and retinoid X receptors: interactions with endogenous retinoic acids. Proc Natl Acad Sci USA 1993; 90:30 - 34
- N dres, J Marill, MA Flexor, GG Chabot. Activation of Retinoic Acid Receptor-dependent Transcription by All-trans-retinoic Acid Metabolites and Isomers. J Biol Chem 2002; 277:31491 - 31498
- Tsukada M, Schroder M, Roos TC, Chandraratna RA, Reichert U, Merk HF, et al. 13-cis retinoic acid exerts its specific activity on human sebocytes through selective intracellular isomerization to all-trans retinoic acid and binding to retinoid acid receptors. J Invest Dermatol 2000; 115:321 - 327
- Schug J, Overton CG. TESS: Transcriptional Element Software Search on the WWW Technical Report CBIL-TR-1997-1001-v0.0 1997; Philadelphia PA Computational Biology and Informatics Laboratory, School of Medicine, University of Pennsylvania
- Hughes BR, Cunliffe WJ. A prospective study of the effect of isotretinoin on the follicular reservoir and sustainable sebum excretion rate in patients with acne. Arch Dermatol 1994; 130:315 - 318
- Stewart ME, Benoit AM, Stranieri AM, Rapini RP, Strauss JS, Downing DT. Effect of oral 13-cis Retinoic Acid at three dose levels on sustainable rates of sebum secretion and on acne. Journal of the American Academy of Dermatology 1983; 8:532 - 538
- Zelickson AS, Strauss JS, Mottaz J. Ultrastructural changes in sebaceous glands following treatment of cystic acne with isotretinoin. Am J Dermatopathol 1986; 8:139 - 143
- Thiboutot D, Jabara S, McAllister J, Sivarajah A, Gilliland K, Cong Z, et al. Human skin is a steroidogenic tissue: Steroidogenic enzymes and cofactors are expressed in epidermis, normal sebocytes, and an immortalized sebocyte cell line (SEB-1). J Invest Dermatol 2003; 120:905 - 914
- Griffiths CE, Russman AN, Majmudar G, Singer RS, Hamilton TA, Voorhees JJ. Restoration of collagen formation in photodamaged human skin by tretinoin (retinoic acid). N Engl J Med 1993; 329:530 - 535
- Kafi R, Kwak HS, Schumacher WE, Cho S, Hanft VN, Hamilton TA, et al. Improvement of naturally aged skin with vitamin A (retinol). Arch Dermatol 2007; 143:606 - 612
- Weiss JS, Ellis CN, Headington JT, Tincoff T, Hamilton TA, Voorhees JJ. Topical tretinoin improves photoaged skin. A double-blind vehicle-controlled study. Jama 1988; 259:527 - 532
- Wang Q, Chen Q, Zhao K, Wang L, Wang L, Traboulsi EI. Update on the molecular genetics of retinitis pigmentosa. Ophthalmic Genet 2001; 22:133 - 154
- Hope WC, Patel BJ, Fiedler-Nagy C, Wittreich BH. Retinoids inhibit phospholipase A2 in human synovial fluid and arachidonic acid release from rat peritoneal macrophages. Inflammation 1990; 14:543 - 559
- Tsukada M, Schroder M, Roos TC, Chandraratna RA, Reichert U, Merk HF, et al. 13-cis retinoic acid exerts its specific activity on human sebocytes through selective intracellular isomerization to all-trans retinoic acid and binding to retinoid acid receptors. J Invest Dermatol 2000; 115:321 - 327
- Chen Y, Kundakovic M, Agis-Balboa RC, Pinna G, Grayson DR. Induction of the reelin promoter by retinoic acid is mediated by Sp1. J Neurochem 2007; 103:650 - 665
- Sitzmann JH, Bauer FW, Cunliffe WJ, Holland DB, Lemotte PK. In situ hybridization analysis of CRABP II expression in sebaceous follicles from 13-cis retinoic acid-treated acne patients. Br J Dermatol 1995; 133:241 - 248
- Eckert RL, Broome AM, Ruse M, Robinson N, Ryan D, Lee K. S100 proteins in the epidermis. J Invest Dermatol 2004; 123:23 - 33
- Lee DY, Yamasaki K, Rudsil J, Zouboulis CC, Park GT, Yang JM, et al. Sebocytes Express Functional Cathelicidin Antimicrobial Peptides and Can Act to Kill Propionibacterium Acnes. J Invest Dermatol 2008;
- Baron JM, Heise R, Blaner WS, Neis M, Joussen S, Dreuw A, et al. Retinoic Acid and its 4-Oxo Metabolites are Functionally Active in Human Skin Cells In Vitro. J Invest Dermatol 2005; 125:143 - 153
- Wolbach SB, Howe PR. Tissue changes following deprivation of fat soluble vitamin A. J Exp Med 1925; 43:753
- Glover JC, Renaud JS, Rijli FM. Retinoic acid and hindbrain patterning. J Neurobiol 2006; 66:705 - 725
- Hoffjan S, Stemmler S. On the role of the epidermal differentiation complex in ichthyosis vulgaris, atopic dermatitis and psoriasis. Br J Dermatol 2007; 157:441 - 449
- Mahajan M, Karmakar S, Weissman S. Control of beta globin genes. Journal of Cellular Biochemistry 2007; 102:801 - 810
- Irizarry RA, Gautier L, Cope LM. The Analysis of Gene Expression Data: Methods and Software Chapter 4 ed 2003a; Spriger Verlag
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, et al. Exploration, normalization and summaries of high density oligonucleotide array probe level data. Biostatistics 2003; 4:249 - 264
- Trivedi NR, Gilliland KL, Zhao W, Liu W, Thiboutot DM. Gene array expression profiling in acne lesions reveals marked upregulation of genes involved in inflammation and matrix remodeling. J Invest Dermatol 2006; 126:1071 - 1079
- Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA 2001; 98:5116 - 5121
- Benjamini Y, Yekutieli D. Quantitative trait Loci analysis using the false discovery rate. Genetics 2005; 171:783 - 790
- Pfaffl MW, Horgan GW, Dempfle L. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res 2002; 30:36
- Wingender E, Dietze P, Karas H, Knuppel R. TRANSFAC: A database on transcription factors and their DNA binding sites. Nucleic Acids Res 1996; 24:238 - 241