Abstract
The aim of our study was to identify relationships between epigenetic parameters correlating with a relaxed chromatin state of the DUX4 promoter region and clinical severity as measured by a clinical severity score or muscle pathologic changes in D4Z4 contraction-dependent (FSHD1) and –independent (FSHD2) facioscapulohumeral muscular dystrophy patients. Twenty primary fibroblast (5 control, 10 FSHD1 and 5 FSHD2) and 26 primary myoblast (9 control, 12 FSHD1 and 5 FSHD2) cultures originating from patients with FSHD and controls were analyzed. Histone modification levels were determined by chromatin immunoprecipitation. We examined correlations between the chromatin compaction score (ChCS) defined by the H3K9me3:H3K4me2 ratio and an age corrected clinical severity score (CSS) or muscle pathology score (MPS). Possible relationships were investigated using linear regression analysis and significance was tested by Pearson’s product-moment coefficient.
We found a significant difference of the ChCS between controls and patients with FSHD1 and between controls and patients with FSHD2. Tissue specific differences in ChCS were also observed. We also found a near-significant relationship between ChCS and the age corrected CSS in fibroblasts but not in myoblasts. Surprisingly, we found a strong correlation between the MPS of the vastus lateralis and the CSS. Our results confirm the D4Z4 chromatin relaxation previously shown to be associated with FSHD in a small number of samples. A possible relationship between clinical and epigenetic parameters could be established in patient fibroblasts, but not in myoblasts. The strong correlation between the MPS of the vastus lateralis and the CSS suggests that this muscle can be used to study for surrogate markers of overall disease severity.
Introduction
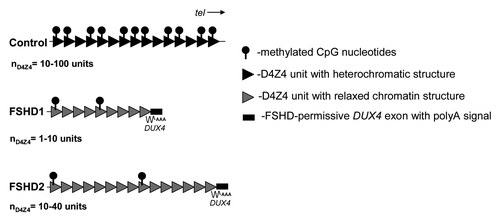
Facioscapulohumeral muscular dystrophy (FSHD) is an autosomal dominant myopathy characterized initially by progressive wasting of the facial, shoulder and upper arm muscles.Citation1 Two distinct groups of affected individuals can be defined: > 95% of patients have a contracted D4Z4 repeat array below 10 units in the subtelomeric region of chromosome 4 (FSHD1) while < 5% of patients do not have a contracted D4Z4 array (FSHD2).Citation2 Both FSHD1 and FSHD2 patients show CpG hypomethylation and specific loss of the repressive histone modification histone 3 lysine 9 trimethylation (H3K9me3) at the D4Z4 array.Citation3,Citation4 While in patients with FSHD1 the hypomethylation is restricted to the contracted allele, both alleles on chromosome 4 and both homologous arrays on chromosome 10 are hypomethylated in FSHD2.Citation5 These epigenetic changes result in a relative chromatin relaxation which may facilitate the expression of full length double homeobox gene 4 (flDUX4) ().Citation4,Citation5 However, in cultured myotubes from patients with FSHD, it is found that flDUX4 expression occurs only on specific genetic backgrounds of chromosome 4 containing an additional DUX4 exon immediately distal to the D4Z4 repeat which stabilizes the transcript and leads to the production of abundant amounts of DUX4 protein in sporadic myonuclei.Citation6
Figure 1. Schematic representation of the FSHD locus in control individuals, patients with contraction dependent (FSHD1) and patients with contraction independent (FSHD2) FSHD. D4Z4 repeat units are indicated with triangles, the additional DUX4 exon with stabilizing polyA signal distal to the repeat is indicated with a box. Common chromatin features of D4Z4 in FSHD1 and FSHD2 are decreased CpG methylation levels and a more relaxed chromatin structure facilitating the expression of DUX4 from the telomeric repeat unit. Overall reduction in CpG methylation of D4Z4 in FSHD is schematically indicated with reduced number of lollipop symbols
The age at onset, severity and disease progression is highly variable between and within FSHD families.Citation1 Several studies addressed the possibility of a correlation between disease severity measured by age at onset or clinical severity score (CSS) and the residual repeat size of the disease-associated D4Z4 repeat in FSHD1 patients.Citation7-Citation9 A rough and inverse correlation was reported between D4Z4 unit number and severity: individuals with 1–3 units typically exhibited the most severe phenotype, while patients with 4–10 D4Z4 units generally show a high variability in clinical presentation.Citation10
Correlations between CpG methylation level of D4Z4 and age-corrected CSS was also studied in FSHD1 and FSHD2 patients. No linear relationship was found between the CpG methylation level of the first D4Z4 unit on the contracted allele in peripheral blood lymphocytes and the age-corrected CSS in a limited number of patients.Citation10
A recent study emphasizes the role of histone modifications in the pathomechanism of FSHD.Citation4 A combined presence of the transcriptionally permissive modification histone 3 lysine 4 dimethylation (H3K4me2) and repressive histone modification H3K9me3 was detected in the promoter region of DUX4. A specific loss of H3K9me3 was reported in immortalized peripheral lymphoblastoid cell lines, primary fibroblast and primary myoblast samples of FSHD patients, both FSHD1 and FSHD2. We hypothesized that the ratio of H3K9me3 and H3K4me2, herein referred to as the chromatin compaction score (ChCS), is significantly different between control and affected individuals and examined the relationship between ChCS and measures of overall disease severity as well as between ChCS and histopathological changes in muscle.
Results
Significant difference in chromatin compaction score between controls and FSHD patients
Since D4Z4 repeats in FSHD1 and FSHD2 patients show identical epigenetic changes, we tested whether the ChCS, the ratio of H3K9me3:H3K4me2, discriminates control samples from FSHD1 and FSHD2 samples. Our study includes 5 control, 10 FSHD1 and 5 FSHD2 primary fibroblast cultures in addition to 9 control, 12 FSHD1 and 5 FSHD2 myoblast cultures. Cross-linked ChIP was performed using H3K9me3 and H3K4me2 antibodies and ChCS were calculated by dividing H3K9me3 relative enrichment values by H3K4me2 relative enrichment values. Statistical analysis of fibroblast data showed significant differences between control and FSHD1 (p < 0.01) and control and FSHD2 samples (p < 0.01) (). ChCS were decreased in patient samples indicating a D4Z4 chromatin relaxation in patient samples.
Figure 2. Scatterplot of ChCS in fibroblasts (A) or myoblasts (B) of controls, FSHD1 and FSHD2 patients. Individual samples are represented by individual data points. Mean values with 1xSD are indicated.
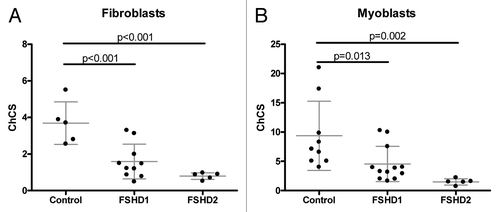
Since FSHD is a progressive muscle disease we hypothesized that a relationship between epigenetic marks and the clinical phenotype is more pronounced in primary myoblast cultures. To avoid possible differences caused by the replicative history of the culture, chromatin was isolated from all samples at passage 6 or 7. Similar to the fibroblast results we found that the ChCS was significantly lower in FSHD1 compared with control (p < 0.05) as well as in FSHD2 compared with control (p < 0.01) samples. We also observed that maximum ChCS values in fibroblast cultures (6 on y axis in ) are lower than maximum ChCS values in myoblast cultures (25 on y axis on ).
Correlation analysis of ChCS and age-corrected CSS in myoblast and fibroblast samples
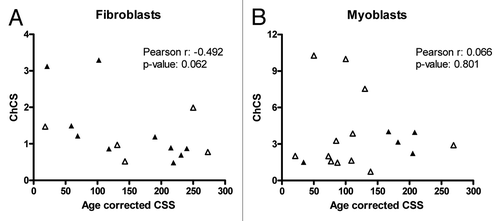
Next we performed linear regression analysis to study the relationship between ChCS and age-correlated CSS. Analysis was done separately on fibroblast and myoblast using all samples and separating them by disease subtype (FSHD1 or FSHD2). Since gender differences have been reported for the penetrance and the severity of the disease, samples were also analyzed after separation based on gender. A summary of analyses with corresponding p values determined by Pearson product moment coefficient are shown in . We found significant negative correlation between ChCS and age-corrected CSS in male FSHD1 and FSHD2 fibroblast samples (p = 0.028) suggesting that the chromatin at D4Z4 is more relaxed in samples originating from males with a more severe phenotype. When combining male and female fibroblast samples the negative correlation reached near-significance (p = 0.062). There was no significant correlation found in myoblasts in any combination of samples studied. Our results of fibroblast (p = 0.062) and myoblast samples (p = 0.801) are presented in .
Table 1. Summary of analyses of age corrected CSS and ChCS
Correlation analysis of ChCS and MPS in myoblast samples
Muscle biopsies were scored for MPS as described in the methods section. Correlation analysis between ChCS and MPS was done on 6 FSHD1 and 5 FSHD2 patients for whom both scores were available (Fig. S1). Statistical analysis showed no relationship between MPS and ChCS.
Correlation analysis of MPS and CSS
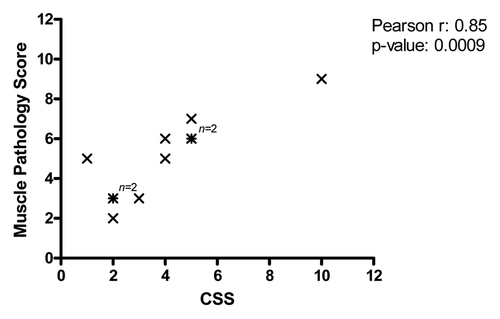
Finally we tested whether we could find a relationship between vastus lateralis MPS and CSS. Since MPS and CSS are compared in the same patient, we refrained from using the age-correction in establishing the clinical severity. As shown in , in the 11 FSHD patients for which both scores were available we found a significant and positive relationship between the muscle pathology score and CSS (r = 0.85; p = 0.0009)
Discussion
FSHD presents with high and unpredictable inter- and intrafamilial clinical variability in disease onset and progression.Citation1 To provide patients with a better prognosis it would be beneficial to identify genetic or epigenetic markers that correlate with disease severity. Earlier studies failed to identify correlations between genetic markers (D4Z4 repeat size) or epigenetic markers (D4Z4 methylation) that can be used in the clinic.Citation8,Citation10
The focus of the present study was to investigate possible relationships between the epigenetic changes occurring in primary fibroblasts and myoblasts of FSHD1 and FSHD2 patients at the promoter region of DUX4 and the clinical severity defined by the CSS or MPS. As a measure of chromatin changes first we introduced the chromatin compaction score, ChCS, in which the relative enrichment of H3K9me3 at D4Z4 is divided by the relative enrichment of H3K4me2. The advantage of such a score is that it circumvents the correlation calculation problem of ChIP experiments studying repetitive sequences. Different samples contain different numbers of D4Z4 units and our experimental procedure detects all repeat units on chromosomes 4 and 10. A comparison between different samples is only reliable if the measured signal can be correlated with D4Z4 unit numbers. Since both H3K9me3 and H3K4me2 are present in every unit, their ratio, as calculated in the ChCS, gives a value per unit.
Statistical analysis revealed a significant difference between the ChCS of control and FSHD1 and between control and FSHD2 in fibroblast and myoblast cultures. These results, showing a significant decrease in ChCS in FSHD cell lines, confirm the previously reported epigenetic alterations at D4Z4 suggesting that the D4Z4 array in FSHD patients is less compact than in controls.Citation4
Comparison of ChCS values between the disease subtypes separately in fibroblast and myoblast samples showed that independent of cell type FSHD1 ChCS values show high variation while FSHD2 ChCS values are lower and less variable. This might be explained by a much more widespread chromatin relaxation over all four repeats in FSHD2, while in FSHD1 the changes may be more restricted to the pathogenic allele. This is also observed for the loss of DNA methylation at D4Z4.Citation5
We also detected tissue-specific differences in the chromatin landscape at D4Z4 arrays of fibroblast and myoblast cultures with control myoblast ChCS values being higher than control fibroblast ChCS values. Genome-wide ChIP-seq studies have shown that the chromatin landscape in a particular cell type reflects its transcriptional activity and is therefore cell type-specific.Citation11 Our results show that ChCS values are also cell-type specific with a more compact chromatin organization of D4Z4 in myoblasts to prevent expression of flDUX4. This may explain the specific tissue involvement in FSHD.
Correlation analysis of the ChCS and age-corrected CSS revealed a significant relationship in male fibroblast patient samples (p = 0.028). Correlation analysis of all fibroblast samples gave borderline significance (p = 0.062). Gender differences for disease penetrance and severity have been reported and showed that males are typically more severely affected than females.Citation12 The weak correlation may be caused by the small sample size or because of technical reasons. In our ChIP assays we measure collectively the chromatin status of each DUX4 promoter in the entire array, rather than specifically interrogating the most telomeric unit from which flDUX4 originates. Moreover, in FSHD1 we are not only measuring the DUX4 promoters of the pathogenic allele, but instead all D4Z4 and D4Z4-like arrays on chromosomes 4 and 10. Considering that in FSHD2 the chromatin status of all four chromosomes seems to be affected, this limitation should not apply to FSHD2. Nevertheless, also in FSHD2 cases we could not detect relationship between ChCS and age corrected CSS possibly due to small sample size.
Statistical analysis of myoblast data could not detect significant relationship between ChCS and age corrected CSS in any combination of samples (). Myoblasts are more sensitive to culturing conditions, often less homogeneous and can quickly transform to a pre-differentiation state, factors that may all affect their epigenetic profile. An alternative explanation might be that myoblast cultures carry a ‘history of disease’ that may affect the chromatin status of D4Z4 while fibroblasts, being derived from a non-affected tissue are not expected to have such history.
Irrespective of the explanation, if a negative correlation between the ChCS and CSS in fibroblast samples can be confirmed in larger group of patients, it would provide the first prognostic marker that can be measured in a relatively accessible tissue. In addition, since we could observe significant ChCS changes in myoblasts and fibroblasts, it is tempting to speculate that this score can also be used for prenatal diagnosis of FSHD2, a condition for which currently no validated diagnostic test exists.
An interesting observation from our study is that there is strong correlation between the CSS and the MPS of the vastus lateralis muscle. These results indicate that the pathology observed in the vastus lateralis muscle, typically not clinically affected in the early stages of FSHD, provides a good reflection of overall disease severity. This result shows that the vastus lateralis muscle, easily and safely accessible by needle biopsy techniques, is an ideal muscle to study for surrogate markers of overall disease severity. A non-invasive modality such as MRI to score the involvement of the vastus lateralis would be even more preferable. A study of lower extremity MRI in FSHD using a simple scoring system based on T1-W sequences was recently completed.Citation13 This study showed that the combined MRI score of all lower extremity muscles was highly correlated with CSS. Whether this correlation holds true when the MRI score is based on the score of only the vastus lateralis remains to be determined.
In summary, our study of a large cohort of patient and control fibroblast and myoblast cultures confirms earlier reports of D4Z4 chromatin relaxation in FSHD and introduces a practical chromatin compaction score (ChCS). We found a near-significant correlation between the ChCS and severity in patient fibroblast samples and identified an unexpected correlation between the CSS and the MPS. We propose that factors in addition to locus intrinsic properties, influence disease severity in FSHD.
Materials and methods
Patients and control individuals
All samples from controls and FSHD patients were obtained after signing informed consents. Samples from 22 FSHD1, 10 FSHD2 and 14 controls were studied. Samples were analyzed for array size, 4q haplotype and DNA methylation according to published methods before the studyCitation3,Citation14 (Tables S1 and S2). Fibroblasts were derived from 2 mm punch skin biopsies obtained from the medial aspect of the distal forearm. Myoblasts were derived from needle muscle biopsy samples taken from the vastus lateralis muscle. An adjacent sample of muscle was processed for histopathology. Muscle and skin biopsy procedures are described here: http://www.urmc.rochester.edu/fields-center/protocols/index.cfm. The muscle pathologic score (MPS) is based on review of four sections stained with Hematoxylin and Eosin as well as modified Gomori Trichrome staining. The 12 point pathologic score (0: normal, 12: end stage muscle) is based on assessment of variability in fiber size, percent of central nuclei, presence of active muscle fiber necrosis and regeneration and amount of fibrofatty replacement (http://dev.mc.rochester.edu/fields-center/protocols/index.cfm).
Cell lines and culturing
All primary myoblast and fibroblast cell cultures were obtained from the University of Rochester’s FSHD biorepository (www.FieldsCenter.org). A list of used primary cell cultures, the 4q35 genotype and methylation status of D4Z4 is shown in Table S1 for myoblast samples and in Table S2 for fibroblast samples.
Myoblast cell lines were cultured in DMEM/F-10 (#31550 GIBCO, Grand Island, NY) supplemented with 20% heat inactivated fetal bovine serum (FBS #10270 GIBCO), 1% penicillin/streptomycin (#15140 GIBCO), 10ng/ml basic rhFGF (#G5071 Promega, Madison, Wisconsin) and 1μM Dexamethasone (#D2915 SIGMA, St. Louis, Missouri).
Fibroblast cell lines were cultured in DMEM/F-12 media supplemented with 20% heat inactivated fetal bovine serum (FBS #10270 GIBCO), 1% penicillin/streptomycin (#15140 GIBCO), 10mM HEPES (#15630 GIBCO), 1mM Sodium Pyruvate (#11360 GIBCO).
Chromatin immunoprecipitation
Chromatin was prepared from cells fixed with 1% formaldehyde according to a published protocol.Citation15 Every sample was independently studied twice. ChIP-grade antibodies against H3K4me2 (no. 39142) and against H3K9me3 (no. 39162) used in this study were purchased from Active Motif (Carlsbad, CA, USA). Normal rabbit serum was used to measure unspecific binding of proteins to beads. Immunopurified DNA was quantified with Q-PCR primer pairCitation4 and quantitative PCR measurements were done with CFX96TM real time system using iQTM SYBRR Green Supermix. Relative enrichment values were calculated by subtracting the IgG ChIP values representing background from the ChIP values with the H3K9me3 or H3K4me2 antibodies.
Statistical analysis
Differences in ChCS between controls, FSHD1 and FSHD2 samples were statistically analyzed by one-way-ANOVA, followed by LSD multiple comparisons test between individual groups. Correlations were tested by linear regression analyses by calculating the Pearson R coefficient with corresponding p-values between the age corrected CSS and ChCS or between CSS and muscle pathology score for different groups of samples.
| Abbreviations: | ||
| FSHD | = | facioscapulohumeral muscular dystrophy |
| ChCS | = | chromatin compaction score |
| CSS | = | clinical severity score |
| ChIP | = | chromatin immunoprecipitation |
| MPS | = | muscle pathology score |
| flDUX4 | = | full length DUX4 |
| H3K4me2 | = | histone 3 lysine 4 dimethylation |
| H3K9me3 | = | histone 3 lysine 9 trimethylation |
Additional material
Download Zip (100.1 KB)Acknowledgments
We would like to thank patients and their families for supporting our research by supplying us with biological material.
Disclosure of Potential Conflicts of Interest
SVDM, SJT and RT are co-inventors on an FSHD patent application.
Financial Disclosure Statement
This work was supported by institutional funds from the Leiden University Medical Center, the Fields Center for FSHD and Neuromuscular Research, the Prinses Beatrix Fonds (WAR08–14), the Netherlands Organization for Scientific Research [NWO 917.56.338], the National Institutes of Health [P01NS069539], the Geraldi Norton Foundation and the Eklund Family.
References
- Padberg GW. Facioscapulohumeral disease. Leiden University, Leiden, 1982.
- de Greef JC, Lemmers RJ, Camaño P, Day JW, Sacconi S, Dunand M, et al. Clinical features of facioscapulohumeral muscular dystrophy 2. Neurology 2010; 75:1548 - 54; http://dx.doi.org/10.1212/WNL.0b013e3181f96175; PMID: 20975055
- van Overveld PG, Lemmers RJ, Sandkuijl LA, Enthoven L, Winokur ST, Bakels F, et al. Hypomethylation of D4Z4 in 4q-linked and non-4q-linked facioscapulohumeral muscular dystrophy. Nat Genet 2003; 35:315 - 7; http://dx.doi.org/10.1038/ng1262; PMID: 14634647
- Zeng W, de Greef JC, Chen YY, Chien R, Kong X, Gregson HC, et al. Specific loss of histone H3 lysine 9 trimethylation and HP1gamma/cohesin binding at D4Z4 repeats is associated with facioscapulohumeral dystrophy (FSHD). PLoS Genet 2009; 5:e1000559; http://dx.doi.org/10.1371/journal.pgen.1000559; PMID: 19593370
- de Greef JC, Lemmers RJ, van Engelen BG, Sacconi S, Venance SL, Frants RR, et al. Common epigenetic changes of D4Z4 in contraction-dependent and contraction-independent FSHD. Hum Mutat 2009; 30:1449 - 59; http://dx.doi.org/10.1002/humu.21091; PMID: 19728363
- Snider L, Asawachaicharn A, Tyler AE, Geng LN, Petek LM, Maves L, et al. RNA transcripts, miRNA-sized fragments and proteins produced from D4Z4 units: new candidates for the pathophysiology of facioscapulohumeral dystrophy. Hum Mol Genet 2009; 18:2414 - 30; http://dx.doi.org/10.1093/hmg/ddp180; PMID: 19359275
- Tawil R, Forrester J, Griggs RC, Mendell J, Kissel J, McDermott M, et al, The FSH-DY Group. Evidence for anticipation and association of deletion size with severity in facioscapulohumeral muscular dystrophy. Ann Neurol 1996; 39:744 - 8; http://dx.doi.org/10.1002/ana.410390610; PMID: 8651646
- Lunt PW, Jardine PE, Koch MC, Maynard J, Osborn M, Williams M, et al. Correlation between fragment size at D4F104S1 and age at onset or at wheelchair use, with a possible generational effect, accounts for much phenotypic variation in 4q35-facioscapulohumeral muscular dystrophy (FSHD). Hum Mol Genet 1995; 4:951 - 8; http://dx.doi.org/10.1093/hmg/4.5.951; PMID: 7633457
- Ricci E, Galluzzi G, Deidda G, Cacurri S, Colantoni L, Merico B, et al. Progress in the molecular diagnosis of facioscapulohumeral muscular dystrophy and correlation between the number of KpnI repeats at the 4q35 locus and clinical phenotype. Ann Neurol 1999; 45:751 - 7; http://dx.doi.org/10.1002/1531-8249(199906)45:6<751::AID-ANA9>3.0.CO;2-M; PMID: 10360767
- van Overveld PG, Enthoven L, Ricci E, Rossi M, Felicetti L, Jeanpierre M, et al. Variable hypomethylation of D4Z4 in facioscapulohumeral muscular dystrophy. Ann Neurol 2005; 58:569 - 76; http://dx.doi.org/10.1002/ana.20625; PMID: 16178028
- Ernst J, Kheradpour P, Mikkelsen TS, Shoresh N, Ward LD, Epstein CB, et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature 2011; 473:43 - 9; http://dx.doi.org/10.1038/nature09906; PMID: 21441907
- Tonini MM, Passos-Bueno MR, Cerqueira A, Matioli SR, Pavanello R, Zatz M. Asymptomatic carriers and gender differences in facioscapulohumeral muscular dystrophy (FSHD). Neuromuscul Disord 2004; 14:33 - 8; http://dx.doi.org/10.1016/j.nmd.2003.07.001; PMID: 14659410
- Frisullo G, Frusciante R, Nociti V, Tasca G, Renna R, Iorio R, et al. CD8(+) T cells in facioscapulohumeral muscular dystrophy patients with inflammatory features at muscle MRI. J Clin Immunol 2011; 31:155 - 66; http://dx.doi.org/10.1007/s10875-010-9474-6; PMID: 21063901
- Lemmers RJ, van der Wielen MJ, Bakker E, Frants RR, van der Maarel SM. Rapid and accurate diagnosis of facioscapulohumeral muscular dystrophy. Neuromuscul Disord 2006; 16:615 - 7, author reply 617-8; http://dx.doi.org/10.1016/j.nmd.2006.07.013; PMID: 16938455
- Nelson JD, Denisenko O, Bomsztyk K. Protocol for the fast chromatin immunoprecipitation (ChIP) method. Nat Protoc 2006; 1:179 - 85; http://dx.doi.org/10.1038/nprot.2006.27; PMID: 17406230