Abstract
In eukaryotes, epigenetic information can be encoded in parental cells through modification of histones and subsequently passed on to daughter cells in a process known as transgenerational epigenetic regulation. Simian Virus 40 (SV40) is a well-characterized virus whose small circular DNA genome is organized into chromatin and, as a consequence, undergoes many of the same biological processes observed in cellular chromatin. In order to determine whether SV40 is capable of transgenerational epigenetic regulation, we have analyzed SV40 chromatin from minichromosomes and virions for the presence of modified histones using various ChIP techniques and correlated these modifications with specific biological effects on the SV40 life cycle. Our results demonstrate that, like its cellular counterpart, SV40 chromatin is capable of passing biologically relevant transgenerational epigenetic information between infections.
Epigenetic regulation through covalent modification of histones has been shown to play a critical role in a number of important biological processes including cell fate determination during differentiation,Citation1-Citation5 regulation of gene expressionCitation6,Citation7 and the development of cancer.Citation8-Citation11 Importantly, all of these examples of epigenetic regulation are characterized by the transfer of epigenetic information from parental cells to daughter cells during cellular division.
Simian Virus 40 (SV40) is a DNA tumor virus that is organized into chromatin in the intracellular minichromosome and virion, and undergoes hyperacetylation of H3 and H4 during transcriptionCitation12-Citation14 and methylation of H4K20 during an infection.Citation15 Because of its similarity to cellular chromatin, we hypothesized that SV40 should be able to transfer epigenetic information recapitulating the conditions of an infection to a subsequent infection through the infecting virions. We tested this hypothesis by characterizing the histone tail modifications in SV40 chromatin derived from minichromosomes and virions using ChIP analyses and correlating the observed variations in histone modification in the SV40 chromatin with subsequent effects on an infection.
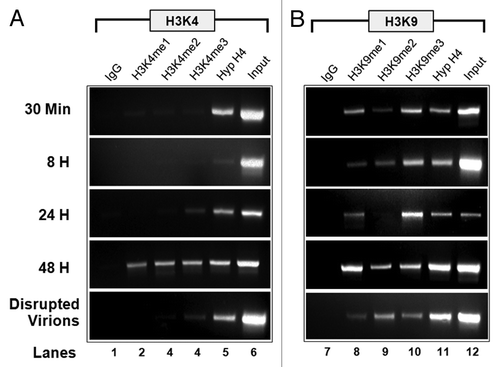
Extending our previous results,Citation15 we analyzed SV40 wild-type chromatin for the presence of methylated H3K4 and H3K9 during an infection, with the results shown in . When SV40 chromatin was analyzed for the presence of methylated H3K4 (), we observed mono, di and trimethylated H3K4 present at 48 h post-infection (PI), but little, if any, methylated H3K4 at other times. When SV40 was analyzed for the presence of methylated H3K9 (), all three of the methylated forms of H3K9 were present in SV40 chromatin throughout the course of the infection, including disrupted virions, with the exception of H3K9me2 at 24 h PI, which was only present at low levels.
Figure 1. Methylated H3K4 (A) and H3K9 (B) are present in SV40 chromatin during a lytic infection. Unfixed SV40 minichromosomes were isolated from disrupted virions and infected cells at 30 min, 8 h, 24 h, and 48 h post-infection (PI), subjected to chromatin immunoprecipitation with 7.5 µl of antibody to each target modified histone, and the resulting immunoprecipitates amplified by PCR with primers recognizing the SV40 early region as previously described.Citation15 (A) Lane 1, IgG; Lane 2, H3K4me1; Lane 3, H3K4me2; Lane 4, H3K4me3; Lane 5 Hyperacetylated H4; Lane 6, 5% of input SV40 chromatin. (B) Lane 1, IgG; Lane 2, H3K9me1; Lane 3. H3K9me2; Lane 4, H3K9me3; Lane 5 Hyperacetylated H4; Lane 6, 5% of input SV40 chromatin. DNA present in immunoprecipitates from virions and minichromosomes isolated 48 h PI was amplified for 32 cycles. DNA from the other immunoprecipitates was amplified for 35 cycles.
If the identified histone modifications actually represented epigenetic information, we expected that the modifications would be organized in the same way as in cellular chromatin, i.e., into various specific combinationsCitation16 that would reflect the biological properties of the chromatin.
SV40 minichromosomes were analyzed by a two-step ChIP process in which the minichromosomes were first immune selected with an antibody to a specific form of modified histone, the chromatin was fragmented, and the resulting fragments subjected to a second ChIP with an antibody to a different modified histone. If two modified forms of histones were found on the same minichromosomes, we would expect to see that a significant fraction of the input chromatin from the first ChIP would be bound in the second ChIP with antibody to the second form of modified histone. shows the result of this analysis.
Table 1. Co-localization of modified histones on SV40 minichromosomes
Based upon these results, we believe that that there are at least five major SV40 epigenomes present in cells infected with wild-type virus at 48 h post-infection. One consisted primarily of H3K4me2 in conjunction with H3K9me1, H3K9me2, H3K9me3, and hyperacetylated H4 (HH4). A second form also consisted primarily of H3K4me2 but in conjunction with HH3 and HH4. This conclusion was based upon the fact that H3K9 cannot be methylated and acetylated at the same time, which we experimentally confirmed. A third epigenome consisted almost solely of H3K4me3 with some HH4 also present. The fourth epigenome consisted of H3K9me1 along with H3K9me2, H4K20me1 and HH4. The fifth epigenome consisted primarily of H3K9me3 with trace amounts of other modifications.
While this is the first demonstration that multiple distinct viral epigenomes defined by specific combinations of histone modifications are present in infected cells, the co-localized modified histones are in agreement with previously reported results for cellular chromatin. For example, H3K9me1 has been previously shown to be co-localized within nucleosomes of transcribing regions with H3K4me1/2/3, H3K79me1/2/3 and H4K20me1 using a genome-wide strategy. H3K9me3 was found associated with H3K9me2 and H3K27me2/3 in silenced regions.Citation16,Citation17
In order to test whether the modified histones in epigenomes present at late times reflected biological properties of the infection, we analyzed wild-type and mutant SV40 minichromosomes defective for the repression of early transcription for the presence of modified histones. The SV40 mutant cs1085 contains a 30 bp deletion of T-antigen binding Site I within the regulatory/promoter region and, as a consequence, does not repress early transcription as seen in a wild-type infection.Citation18,Citation19 This deletion results in an over-production of early mRNA and protein. We have previously exploited this aspect of cs1085 to investigate nucleosome phasingCitation20 and RNA polymerase II occupancy in transcribing SV40 minichromosomes.Citation12 SV40 early expression is also regulated by a miRNA found in the late region of the genome.Citation21 A mutant, SM (SV40 miRNA mutant), deleted for the regulatory miRNA without affecting any other aspects of the virus, was shown to result in the over-production of T-antigen during infection.Citation21
We hypothesized that if one or more of the histone modifications observed in the epigenomes at 48 h PI in a wild-type infection was the result of T-antigen binding to Site I and subsequent repression of early transcription, these modified histones would likely be absent or significantly reduced in cs1085 minichromosomes late in infection. Alternatively, if any of the histone modifications were a result of overexpression of T-antigen one might expect similar effects by both mutants.
SV40 minichromosomes were prepared at 48 h PI from cells infected with wild-type, cs1085, or SM virus and analyzed for the presence of methylated H3K4 and H3K9. The levels of the various modified histones in each of the minichromosomes were then quantitated by real time PCR and compared as shown in . In the absence of T-antigen mediated repression in cs1085 infections, we observed the greatest reduction in the amount of H3K9me1, and significant reductions in the amount of H3K4me1 and H3K4me2. In contrast, in the minichromosomes from SM virus infections with over-production of T-antigen, we observed increases in the amounts of H3K9me1, H3K9me2 and H3K9me3. These results indicated that T-antigen mediated repression of early transcription had its primary effect on the introduction of H3K9me1, a modification thought to be associated with repression in other eukaryotes.Citation22 Unexpectedly, T-antigen mediated repression also appeared to have an effect on the introduction of H3K4me1 and H3K4me2 into SV40 minichromosomes, modifications generally thought to be associated with transcriptional activation.Citation22 Based upon these results, it would appear that the epigenome containing large amounts of H3K9me1 along with H3K4me1/2 was mostly likely associated with repression of early transcription.
Table 2A. Presence of modified histones in SV40 minichromosomes
We then directly tested the hypothesis that SV40 virions could carry epigenetic information by analyzing the histone modifications present in chromatin from virions. If the virions carried epigenetic information one would expect the patterns of histone modifications in the virions to relate to the patterns present in the minichromosomes. In contrast, if the pattern present in the virions was a result of other biological processes, one might expect the pattern in the virions to be independent of the patterns present in the minichromosomes. Virions were purified, disrupted with DTT, the chromatin present in the disrupted virions purified from intact virions,Citation15 and analyzed for the presence of modified histones with the results shown in . Compared with the pattern of H3K9 methylation and HH4 acetylation in wild-type chromatin from virions, we observed a significant reduction in the percentage of chromatin containing the three methylated forms of H3K9 and HH4 in cs1085 and a significant reduction in the percentage of H3K9me2, H3K9me3 and HH4 in the SM mutant. While the pattern of changes in the virions closely mirrored the pattern of histone modifications in the cs1085 minichromosomes, the pattern for the chromatin in SM virions did. Nevertheless, both mutants displayed patterns of histone modifications in the virions that were very different from the wild-type virion chromatin and each other, indicating that in each case the virion chromatin carried distinct epigenetic information.
Table 2B. Presence of modified histones in SV40 virions
Next, we confirmed that the pattern of histone modifications in virion chromatin reflected aspects of the infection by comparing virions produced at 37°C and 39°C. Since the SV40 DNA sequence is identical at both temperatures, any changes in the pattern of histone modifications in the virions must be a result of epigenetic changes. As shown in the pattern of histone modifications for virions produced at 39°C was distinct and differed from the results obtained from the wild-type virions and both mutants particularly with respect to the amount of H3K9me3. These results were consistent with the hypothesis that the conditions of an infection contribute to the epigenetic modifications appearing in the virions produced during that infection.
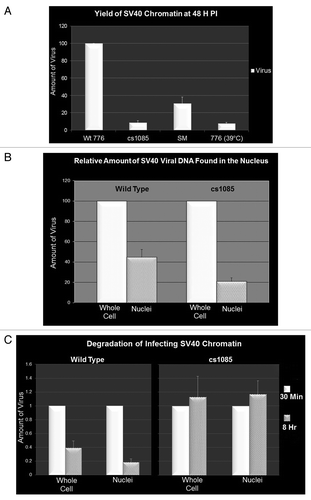
If the patterns of modified histones present in chromatin from virions represented inheritable epigenetic information, we expected that this epigenetic information would affect aspects of a subsequent infection. In order to test this hypothesis, we analyzed infections by the various viruses for the yield of SV40 DNA at 48 h PI as a measure of infection efficiency. SV40 DNA in virions and infected cells was isolated by the Hirt procedureCitation23 and measured by real time PCR. All comparative infections contained the same amount of input SV40 DNA in the pool of infecting virus. Consistent with our hypothesis that the viruses contained inheritable epigenetic information, in comparison to the wild-type virus prepared at 37°C, whose amount was set at 100%, we observed only 9 ± 2% for cs1085, 31 ± 7% for SM and 8 ± 1% for the virus prepared at 39°C ().
Figure 2. Methylation of histone H3 affects infectivity and stability of infecting SV40 chromatin. Cells were infected with equal amounts (based upon real time quantitation of the SV40 DNA present in virions) of wild-type 776, cs1085, SM or 776 grown at 39°C virus. SV40 DNA or minichromosomes were isolated at the times indicated and quantitated by real time PCR. (A) Yield of SV40 DNA at 48 h PI following infection with wild-type 776, cs1085, SM virus or 776 grown at 39°C. (B) SV40 DNA present in cells or nuclei infected with equal amounts of 776 or cs1085 virus at 30 min PI. C. SV40 DNA present in cells infected with equal amounts of 776 or cs1085 virus at 30 min and 8 h PI.
Since the epigenetic information encoded in the histone modifications present in virion chromatin is first made available for reading following uncoating of the virus particle, we hypothesized that this epigenetic information might have an effect on the early stages of the establishment of an infection. In order for an SV40 infection to be established, virions must be transported through the cytoplasm, presented to the nuclear pore complex, and the protein coat of the virion removed or substantially modified so that cellular factors can access the chromatin to initiate early transcription.Citation24,Citation25 In a competing process, we have previously reported that a significant fraction of incoming viral chromatin is targeted for degradation by the cell.Citation15
In order to test whether the observed differences in wild-type and cs1085 epigenetic information affected transport to the nucleus or cellular degradation of viral chromatin, we measured by real time PCR the amount of SV40 DNA present as chromatin in whole cells and nuclei at 30 min and 8 h following infection with the two SV40 viruses. For these studies, nuclei were prepared according to our standard protocol, which was originally optimized for the yield of biologically active SV40 minichromosomes.Citation26 As shown in , a significantly smaller percentage of infecting SV40 chromatin was found in the nucleus following infection with cs1085 compared with wild-type virus at 30 min PI indicating that the epigenetic information carried by the two viruses had an effect on import into the nucleus. The epigenetic information carried by the infecting virions also had an effect on cellular degradation of the SV40 chromatin. As shown in , wild-type chromatin was degraded approximately 75% between 30 min and 8 h when measured in either whole cells or nuclei, consistent with our previous results,Citation15 while cs1085 chromatin appeared to be resistant to degradation (). These results indicate that both the passage through the nuclear pore complex and the degradation of the incoming viral chromatin are sensitive to the specific epigenetic information carried by the viral chromatin.
While this first demonstration of virion-dependent transfer of biologically relevant epigenetic information between infections utilized SV40, which is generally thought to be nonpathogenic in humans, it is very likely that other DNA viruses that are organized into chromatin and are pathogenic (e.g., papillomaviruses) will also show this behavior. For those viruses that utilize epigenetics in the establishment of an infection, these results suggest that this process is much more complex and more highly regulated than previously thought. Finally, the results with SV40 virions prepared at elevated temperature indicate that fevers during viral infection may affect the disease process through the disruption of the normal epigenetic regulation pathways of the viral life cycle.
Materials and Methods
Cells, viruses and infections
SV40 DNA and minichromosomes were prepared in the BSC-1 cell line (ATCC) using wild-type 776 (from Dr. Daniel Nathans), cs1085 (from Dr. Daniel Nathans) or SM virus (from Dr. Chris Sullivan). Uninfected cells were maintained at 37°C in minimal essential medium containing 10% (v/v) fetal calf serum, gentamycin, glutamine, and sodium bicarbonate (GIBCO) as previously described.Citation12-Citation14 Sub-confluent cell mono-layers were exposed to virus for 30 min, the virus containing medium removed, the plates washed twice with fresh warm medium, 10 ml of fresh medium added, and the infected cells incubated at 37°C for the times indicated as described previously.Citation12-Citation14 SV40 minichromosomes were prepared in 75 cm2 T- flasks containing a total of 10 ml of medium, while experiments in which SV40 DNA was prepared were performed in 25 cm2 T-flasks containing 3 ml of medium. Comparative infections with either different viruses or for different periods of infection were performed in parallel at the same time.
Preparation of SV40 DNA, nuclei and minichromosomes
Total SV40 DNA present in infected cells was prepared according to the Hirt procedure.Citation23 Briefly, the medium was removed from the infected cells, the cells washed twice with cold PBS, and 0.3 ml of lysing solution (0.6% SDS, 10 mM EDTA) added. The flask was rocked for a few minutes until all the cells in the flask appear to be solubilized and the solution transferred to a 1.4 ml Eppendorf tube. Following transfer the flask was washed with 0.2 ml of PBS and the wash added to the 0.3 ml transferred to the tube. 160 µl of 5M NaCl was added to the tube and the tube inverted 20 times to mix the contents. After storage overnight at 4°C, the tube was spun at 16,000 x G at 4°C for 30 min, and the soluble contents removed from the cellular DNA pellet. The soluble fraction was referred to as the Hirt supernatant and was used in subsequent analyses.
The SV40 DNA present in the nuclei of infected cells was prepared from infected cells (25 cm2 T-flasks) by a modification of our previously described procedure for preparing SV40 minichromosomes. Nuclei were released from infected cells by a combination of scraping the cells in low-ionic strength buffer (0.5 ml) and TritonX-100 (0.5 ml) as previously describedCitation12-Citation14 and the liquid transferred to a 1.4 ml Eppendorf tube. Any remaining cells in the flask were obtained by a second scraping treatment in low-ionic strength buffer (0.5 ml) and TritonX-100 (0.5 ml) and transfer of the liquid to a second Eppendorf tube. The tubes were spun at 16,000 x G for 30 sec and the supernatant removed. The pellet in each tube was suspended in 0.2 ml of low-ionic-strength buffer, the supernatants combined and gently layered onto a new tube containing 0.6 ml of low-ionic-strength buffer. Following centrifugation at 5000 x G for 10 min, then supernatant was removed and the pellet resuspended in 0.2 ml of PBS. Lysing solution (0.3 ml) was added followed by 0.16 ml of 5M NaCl. The tube was inverted about 20 times and placed overnight at 4°C. The next day the tube was spun at 16,000 x G for 30 min at 4°C in an Eppendorf Model 5415 C centrifuge. The supernatant was transferred to a new tube labeled nuclear portion and the pellet was discarded. SV40 minichromosomes were prepared from infected cells and purified on glycerol step gradients as previously described.Citation12-Citation14 Gradient fractions 3 through 5 were pooled for subsequent analyses.
Preparation of SV40 chromatin from virions
SV40 virus was prepared by infecting cells with 0.005 plaque-forming units of seed virus per cell in order to limit the amount of defective virus formed. Infected cells were incubated until approximately 99% of the cells were dead. Crude virus (1 ml) was transferred to a 1.4 ml Eppendorf tube and centrifuged in a Beckman TLA 100 ultracentrifuge at 50,000 x G for 35 min at 4°C on 10% glycerol buffer (10 mM HEPES, 5 mM KCl, 1mM EDTA, 0.2 mM MgCl2, 0.5 mM DTT, 10% glycerol). The pellet of virus and cellular debris was resuspended in 175 µl of TE buffer, and 20 µl of a 10X digestion buffer supplied by NEB added and 5 µl of DNaseI (NEB) added. Following incubation at 37°C for 15 min to allow the DNase I to degrade any non-virion forms of SV40 chromatin, the mixture was centrifuged as above in the TLA 100 ultracentrifuge on 10% glycerol buffer. The pellet was resuspended in 194 µl of TE buffer, 6 µl of 5 M DTT was added, and the suspension incubated at room temperature for 15 min to disrupt the virions as previously described.Citation15 The mixture was again sedimented on 10% glycerol as described above. Fractions 2 through 5 which have previously been shown to contain SV40 chromosomes were pooled and used in subsequent analyses.
Standard chromatin immunoprecipitation analyses
All of the antibodies used in these studies were ChIP validated by the supplier and included H3K4me1 (07–436, Millipore), H3K4me2 (39141, Active Motif), H3K4me3 (04–745, Millipore), H3K9me1 (ab9045, Abcam), H3K9me2 (ab1220, Abcam), H3K9me3 (ab8898, Abcam), H4K20me1 (39175, Active Motif), hyperacetylated H3 (06–599, Millipore), and hyperacetylated H4 (06–866 Millipore). We also used an antibody recognizing H4K20me1 which was a gift from Dr. Thomas Jenuwein, which we previously showed functioned similarly to the commercial antibody.Citation15 Unfixed SV40 chromatin (100 µl) from infections and disrupted virions was immunoprecipitated with 10 µl of antibody using the reagents and protocol supplied by Millipore in their kit for the analysis of hyperacetylated histone H4 with minor modifications.Citation12-Citation14
Preparation of DNA for PCR analyses
DNA samples for standard PCR were prepared by phenol/chloroform extraction followed by ethanol precipitation in the presence of paint pellet (Novagen) as previously described.Citation12-Citation14 DNA samples for real time PCR were prepared by phenol/chloroform extraction and used directly in amplifications. DNA samples from ChIP elutions (10 µl) were mixed with 50 µl of phenol/chloroform (AM9730, Ambion) and TE buffer (70 µl). The mixture was vortexed for 10 sec and centrifuged for 10 min at 8,000 x G in an IEC Micromax centrifuge. The aqueous phase (40 µl) was removed for subsequent PCR amplification.
PCR amplifications
Purified DNA samples were amplified by standard PCR using primers which recognize the early region of the SV40 genome (5′GCTCCCATTCATCAGTTCCS3′ and 5′CTGACTTTGGAGGCTTCTGG3′) as previously described.Citation12-Citation14 Amplification was initiated with a hot start at 95°C for 2 min followed by 35 to 45 cycles of amplification. Each cycle consisted of an annealing step at 60°C for 1 min, DNA syntheses at 72°C for 1 min, and denaturation at 95°C for 1 min.
Real time PCR
SV40 DNA was amplified from the early region of the SV40 genome using the primers described above (0.3 µM) in a Bio-Rad MyIQ2 real time thermal cycler using the Power SYBr Green PCR master mix (Applied Biosystems). The amplifications were initiated by a hot start at 94°C for 15 min. The DNA was amplified for between 40 and 50 cycles with each cycle consisting of annealing of primers at 60°C for 1 min, DNA synthesis at 72°C for 1 min, and denaturation of the DNA at 94°C for I min. All samples were analyzed in duplicate sets.
Analysis of PCR amplification products
Following standard PCR amplifications, amplification products were separated on 2.4% submerged agarose gels (Sigma) by electrophoresis.Citation12-Citation14 Amplification products were visualized by staining with ethidium bromide and digitally recorded using a UVP GDS8000 gel documentation system (Ultra Violet Products).
Acknowledgments
This work was supported by grants from the National Institutes of Health GM074811 and AI070193 to BM
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Author Contributions
B.M and L.B jointly conceived the studies with input from L.K for the in vivo studies. B.M., L.K, A.G., N.A. and E.W performed the experiments. B.M and L.B. prepared the manuscript. All authors read, edited, and approved the final manuscript.
References
- Atkinson SP, Koch CM, Clelland GK, Willcox S, Fowler JC, Stewart R, et al. Epigenetic marking prepares the human HOXA cluster for activation during differentiation of pluripotent cells. Stem Cells 2008; 26:1174 - 85; http://dx.doi.org/10.1634/stemcells.2007-0497; PMID: 18292213
- Corry GN, Tanasijevic B, Barry ER, Krueger W, Rasmussen TP. Epigenetic regulatory mechanisms during preimplantation development. Birth Defects Res C Embryo Today 2009; 87:297 - 313; http://dx.doi.org/10.1002/bdrc.20165; PMID: 19960551
- Lister R, Pelizzola M, Kida YS, Hawkins RD, Nery JR, Hon G, et al. Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stem cells. Nature 2011; 471:68 - 73; http://dx.doi.org/10.1038/nature09798; PMID: 21289626
- Shafa M, Krawetz R, Rancourt DE. Returning to the stem state: epigenetics of recapitulating pre-differentiation chromatin structure. Bioessays 2010; 32:791 - 9; http://dx.doi.org/10.1002/bies.201000033; PMID: 20652894
- Skinner MK. Environmental epigenetic transgenerational inheritance and somatic epigenetic mitotic stability. Epigenetics 2011; 6:838 - 42; http://dx.doi.org/10.4161/epi.6.7.16537; PMID: 21637037
- Bonasio R, Tu S, Reinberg D. Molecular signals of epigenetic states. Science 2010; 330:612 - 6; http://dx.doi.org/10.1126/science.1191078; PMID: 21030644
- Gibney ER, Nolan CM. Epigenetics and gene expression. Heredity (Edinb) 2010; 105:4 - 13; http://dx.doi.org/10.1038/hdy.2010.54; PMID: 20461105
- Herceg Z. Epigenetic information in chromatin and cancer. Eur J Cancer 2009; 45:Suppl 1 442 - 4; http://dx.doi.org/10.1016/S0959-8049(09)70082-6; PMID: 19775664
- Kubicek S, Schotta G, Lachner M, Sengupta R, Kohlmaier A, Perez-Burgos L, et al. The role of histone modifications in epigenetic transitions during normal and perturbed development. Ernst Schering Res Found Workshop 2006; 57:1 - 27; http://dx.doi.org/10.1007/3-540-37633-X_1; PMID: 16568946
- Sawan C, Herceg Z. Histone modifications and cancer. Adv Genet 2010; 70:57 - 85; http://dx.doi.org/10.1016/B978-0-12-380866-0.60003-4; PMID: 20920745
- Uribe-Lewis S, Woodfine K, Stojic L, Murrell A. Molecular mechanisms of genomic imprinting and clinical implications for cancer. Expert Rev Mol Med 2011; 13:e2; http://dx.doi.org/10.1017/S1462399410001717; PMID: 21262060
- Balakrishnan L, Milavetz B. Reorganization of RNA polymerase II on the SV40 genome occurs coordinately with the early to late transcriptional switch. Virology 2006; 345:31 - 43; http://dx.doi.org/10.1016/j.virol.2005.09.039; PMID: 16242748
- Balakrishnan L, Milavetz B. Histone hyperacetylation during SV40 transcription is regulated by p300 and RNA polymerase II translocation. J Mol Biol 2007; 371:1022 - 37; http://dx.doi.org/10.1016/j.jmb.2007.06.080; PMID: 17658552
- Balakrishnan L, Milavetz B. Histone hyperacetylation in the coding region of chromatin undergoing transcription in SV40 minichromosomes is a dynamic process regulated directly by the presence of RNA polymerase II. J Mol Biol 2007; 365:18 - 30; http://dx.doi.org/10.1016/j.jmb.2006.09.044; PMID: 17055528
- Balakrishnan L, Gefroh A, Milavetz B. Histone H4 lysine 20 mono- and tri-methylation define distinct biological processes in SV40 minichromosomes. Cell Cycle 2010; 9:1320 - 32; http://dx.doi.org/10.4161/cc.9.7.11123; PMID: 20404477
- Jenuwein T, Allis CD. Translating the histone code. Science 2001; 293:1074 - 80; http://dx.doi.org/10.1126/science.1063127; PMID: 11498575
- Wang Z, Zang C, Rosenfeld JA, Schones DE, Barski A, Cuddapah S, et al. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet 2008; 40:897 - 903; http://dx.doi.org/10.1038/ng.154; PMID: 18552846
- DiMaio D, Nathans D. Cold-sensitive regulatory mutants of simian virus 40. J Mol Biol 1980; 140:129 - 42; http://dx.doi.org/10.1016/0022-2836(80)90359-9; PMID: 6251230
- DiMaio D, Nathans D. Regulatory mutants of simian virus 40. Effect of mutations at a T antigen binding site on DNA replication and expression of viral genes. J Mol Biol 1982; 156:531 - 48; http://dx.doi.org/10.1016/0022-2836(82)90265-0; PMID: 6288959
- Kube D, Milavetz B. Generation of a nucleosome-free promoter region in SV40 does not require T-antigen binding to site I. Virology 1989; 172:100 - 5; http://dx.doi.org/10.1016/0042-6822(89)90111-6; PMID: 2549706
- Sullivan CS, Grundhoff AT, Tevethia S, Pipas JM, Ganem D. SV40-encoded microRNAs regulate viral gene expression and reduce susceptibility to cytotoxic T cells. Nature 2005; 435:682 - 6; http://dx.doi.org/10.1038/nature03576; PMID: 15931223
- Kouzarides T. Chromatin modifications and their function. Cell 2007; 128:693 - 705; http://dx.doi.org/10.1016/j.cell.2007.02.005; PMID: 17320507
- Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol 1967; 26:365 - 9; http://dx.doi.org/10.1016/0022-2836(67)90307-5; PMID: 4291934
- Cohen SAS, Au S, Panté N. How viruses access the nucleus. Biochim Biophys Acta 2011; 1813:1634 - 45; http://dx.doi.org/10.1016/j.bbamcr.2010.12.009; PMID: 21167871
- Greber UF, Kasamatsu H. Nuclear targeting of SV40 and adenovirus. Trends Cell Biol 1996; 6:189 - 95; http://dx.doi.org/10.1016/0962-8924(96)10016-7; PMID: 15157471
- Tsubota Y, Waqar MA, Burke JF, Milavetz BI, Evans MJ, Kowalski D, et al. Association of enzymes with replicating and nonreplicating simian virus 40 chromosomes. Cold Spring Harb Symp Quant Biol 1979; 43:693 - 704; http://dx.doi.org/10.1101/SQB.1979.043.01.077; PMID: 226313