Abstract
Human Cytomegalovirus (HCMV) is a ubiquitous herpesvirus that infects and establishes latency in the majority of the human population and may cause fatal infections in immunocompromised patients. Recent data implies a close interaction between HCMV encoded proteins and cellular epigenetic mechanisms such as histone acetylation and deacetylation. In this study, we investigated the interactions between HCMV infection and the DNA methylation machinery in different host cells using several approaches. We found that colon cancer cell line HCT-116 lacking the DNMT1 and DNMT3b methyltransferases was susceptible to HCMV-AD169 infection, while wild-type cells were non-susceptible. Treatment of wild-type HCT-116 cells with 5-azacytidine rendered them susceptible to infection. Further investigation of HCMV infected MRC-5 fibroblasts demonstrated significant global hypomethylation, a phenomenon that was virus strain-specific and associated with the re-localization of DNMT1 and DNMT3b from the nucleus to the cytoplasm. The cytoplasmic accumulation of DNMT1 was also evident in in vitro infected macrophages and in epithelial cells in tissue samples from patients with inflammatory bowel disease and concomitant HCMV infection. Foscavir treatment of virus infected fibroblasts did not affect the majority of the virus induced nuclear exclusion of DNMT1, which suggest that it is dependent on viral IE gene products. In conclusion, HCMV infection results in profound effects on the host cell DNA methylation machinery and is associated with inflammation in vivo. Our results improve the understanding of cytomegalovirus pathogenesis and open the search for new antiviral therapy targets. These findings may also contribute to the further understanding of mechanisms involved in DNA methylation abnormalities in physiological and pathological conditions.
Introduction
Human cytomegalovirus (HCMV) is a member of the herpes virus family. It causes a widespread, persistent infection in a majority of the human population. HCMV normally remains dormant after a primary infection in immunocompetent individuals but can cause life-threatening disease in immunocompromised individuals, such as transplant patients, AIDS patients, and congenitally infected neonates.Citation1,Citation2
Reactivated infection in immunocompromised individuals is often associated with inflammation and has highlighted the possible role of HCMV in inflammatory diseases and cancer. HCMV infection has been reported as a risk factor in several inflammatory diseases including inflammatory bowel disease (IBD), systemic lupus erythematosus (SLE) and rheumatoid arthritis as well as in cardiovascular diseases.Citation3,Citation4,Citation8,Citation43,Citation44 An active infection was also reported in several human malignancies, including glioblastoma multiformeCitation5 and in cancers of epithelial origin in organs such as the prostate, breast and colon.Citation6,Citation7,Citation45 Importantly, healthy tissues in close proximity to these tumors remain HCMV negative.
HCMV is not considered to be oncogenic; instead, the virus may affect cancer development by possessing oncomodulatory properties.Citation9 These include specific mechanisms that control cellular and immunological functions that may have an impact on cancer development, e.g., induced telomerase activity,Citation10 control of cellular differentiation, proliferation induced migration/invasiveness and angiogenesis, basic epigenetic functions and immune evasion strategies.Citation11
Molecular characterization of HCMV strains predicts that the 220–240 kb double stranded DNA viral genome has 252 open reading frames; it is believed to encode about 170 proteins, but only 45 to 57 of these genes are essential for viral replication. Hence, the vast majority of viral proteins are involved in modulation of virus-host interactions,Citation12 and may contribute to inflammation and cancer.Citation11
The genomes of herpes viruses, including HCMV, exist as episomes during infection. The episomes are associated with nucleosomes in the infected host cells, but when encapsidated into virions, the viral DNA lacks histones.Citation13,Citation46 It was reported that the host cell nucleosome deposition machinery targets HCMV DNA upon infection, resulting in a stepwise and dynamic viral-chromatin assembly process.Citation14 Based on these findings, it was suggested that epigenetic events are involved in all viral DNA based processes during HCMV infection including genome replication, DNA damage response and the temporal cascade of viral gene transcription.Citation14 In addition, an in vitro model of HCMV latency and reactivation in dendritic cells implies an important role of chromatin remodeling during reactivation of latent virus.Citation15 Several other studies also demonstrate that HCMV gene expression is controlled by epigenetic mechanisms such as histone modifications and DNA methylation.Citation16,Citation17 DNA methylation was also recently shown to be involved in the control of the HIV-1 latency.Citation18 Thus, viral control of epigenetic mechanisms appears to be pivotal for the life cycle of viruses (reviewed in ref. Citation47).
Epigenetics is generally defined as the dynamic and cellularly heritable properties of genome function, influenced by DNA methylation and post-translational modifications of the N-terminal tails of core histones, which participate in regulating gene expression without changing the primary DNA sequence (Reviewed inCitation19). In vertebrates, DNA methylation is mediated by DNA methyltransferase enzymes (DNMTs), which transfer a methyl group from S-adenosylmethionine to the C-5 position of cytosine residues present in CpG dinucleotide sequences. There are currently three known DNA methyltransferases with catalytic activity: DNMT1, DNMT3a and DNMT3b. DNMT1 is considered as a maintenance enzyme since it has high affinity for hemimethylated DNA during DNA replication. Although substantial overlap exists, DNMT3a and DNMT3b are considered primarily de novo methyl transferases, due to their role in methylation of unmethylated DNA sequences.Citation20 DNA methylation is an important player in early development and cancer, and genome-wide hypomethylation and local hypermethylation of tumor suppressor genes are common features of cancer cells.Citation21 Abnormal DNA methylation is also found in other types of disease. Global DNA hypomethylation was found in SLE and other autoimmune diseases.Citation22 Global DNA hypermethylation and gene specific methylation has been implicated in cardiovascular diseases.Citation23,Citation24 Although DNA methylation changes in diseases are well established by numerous studies, underlying mechanistic explanations are not yet completely defined.Citation25
In this study, we investigated the impact of HCMV infection on the host cell DNA methylation machinery. We show that the methylation state influences HCMV infection capacity in vitro. We also demonstrate that HCMV infection in susceptible cells alters the intracellular localization of DNMTs in vitro and in vivo, which was associated with profound reduced global DNA methylation capacity. These observations give further mechanistic insights into HCMV pathogenesis and suggest new targets of antiviral therapy.
Results
DNMT expression influences the susceptibility to HCMV infection
We investigated whether limitation of the DNA methylation machinery affects HCMV replication. The wild-type HCT-116 colon carcinoma cell line, and three knockout cell lines in which DNMT1, DNMT3b or both DNMT1/DNMT3b are knocked out were used. The double knockout cell line has a very low level of global DNA methylation.Citation26,Citation27 We infected all four cell lines with the HCMV laboratory strain AD169 or the HCMV endothelial cell adapted clinical isolate VR1814 and the expression of HCMV IE proteins was evaluated by immunostaining at 3 d post infection (dpi) (). While wild-type HCT-116 cells were non-susceptible to infection by the laboratory strain AD169, HCT-DNMT1KO and HCT-DNMT3bKO cells displayed partial susceptibility to AD169 infection. Interestingly, the HCTDKO cells were shown to be substantially more susceptible to infection with AD169 than HCT-DNMT1KO and HCT-DNMT3bKO cells, as evident by the increased number of infected cells, indicating that the methylation capacity influences HCMV infection. All cell lines were highly susceptible to the endothelial cell adapted clinical isolate VR1814 suggesting that this strain possesses additional mechanism(s) to overcome the inhibitory effect of DNA methylation enzymes.
Table 1. Infection of DNMT knockout HCT-116 cell lines by HCMV strains VR1814 and AD169.
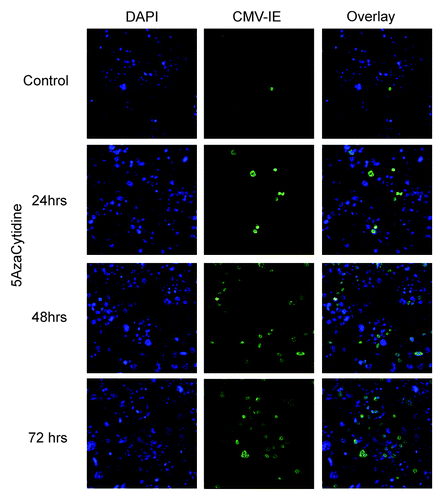
To further explore the importance of the DNA methylation state during HCMV infection, HCT-116 wild-type cells were treated with the DNA methylation inhibitor 5-azacytidine. This treatment turned the cells into a susceptible state and subsequent infection by AD169. The effect of 5-azacytidine was time dependent and reached a maximum after 3 d of treatment (), which indicates that the 5-azacytidine effect is DNA replication dependent, as previously discussed in reference Citation28.
HCMV infection causes DNA hypomethylation
The requirements of a demethylated host cell state for the AD169, but not the VR1814 strain, led us to examine whether the VR1814 strain has the capacity to alter the host cell DNA methylation machinery. We infected MRC-5 cells with HCMV-VR1814 and assayed the global methylation level of the cells at 3 dpi. Analysis of DNA extracted from MRC-5 cells infected with VR1814 revealed a profound hypomethylation compared with uninfected cells. This was evident by two different assays measuring global DNA methylation levels: LUMA and NNA (). The LUMA assay revealed an approximately 85% decrease (from 30% to less than 5%) in global DNA methylation at 3 dpi by VR1814 (), while NNA showed an approximately 45% (from 75% to 40%) reduction in global DNA methylation (), as compared with uninfected cells. Infection with the AD169 strain resulted in less hypomethylation, where the numbers were 10% and 8% respectively. However, neither LUMA nor NNA assays give sequence-specific methylation information, and cannot discriminate whether DNA methylation is decreased in the host genome or if newly replicated viral genomes are prevented from becoming methylated. To address this, we treated HCMV-VR1814 infected MRC-5 cells with Foscavir to inhibit viral DNA replication. By this treatment, no change in DNA methylation was observed between uninfected and infected cells (data not shown), which implies that the main source of hypomethylated DNA in infected cells originates from the newly replicated viral genome.
Figure 2. HCMV infection results in decreased DNA methylation. MRC-5 fibroblasts were infected with HCMV-AD169 or -VR1814. DNA methylation was measured after 3 dpi by Luminometric methylation assay (A) and nearest neighbor analysis (B). Bars represent the mean values. LUMA was repeated 4 times. Error bars denote ± standard deviation and p-values calculated by Student’s t-test. ** = p < 0.01
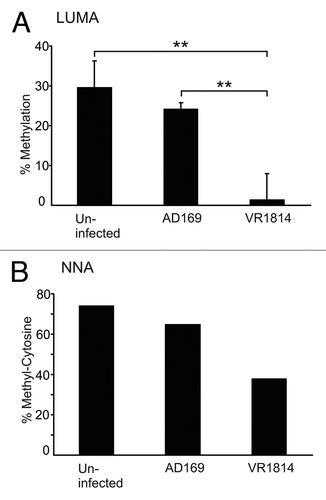
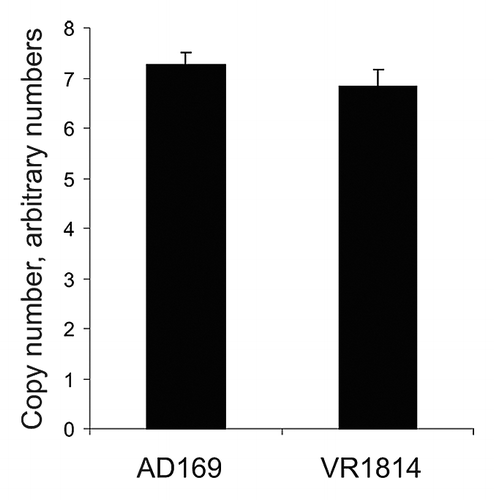
To further investigate the source of the hypomethylated DNA, we interrogated the human-specific methylome in MRC-5 cells, after both HCMV-VR1814 and AD169 infection, using the Illumina Infinium 27K bead chip analysis. However, no specific alterations in DNA methylation were observed in the respective infected cells (Fig. S1). Since the hypomethylation may be due to newly replicated viral DNA, we wanted to assess whether the more profound DNA hypomethylation in VR1814 infected cells as compared with AD169 infected cells was due to strain-specific content of intracellular viral DNA. We therefore performed quantitative real time PCR of the HCMV genome using primer for the HCMV gene pp150.Citation29 The results show that VR1814 and AD169 genomes are present in similar amounts when cells were infected with an MOI of 5 (). Since the apparent methylation levels were different between the two viral strains, we also assessed the strain-specific methylation in a direct way by using methylated DNA Immunoprecipitation (MeDIP) of DNA from infected MRC-5 cells, followed by virus-specific qRT-PCR using primers specific to the HCMV DNA Polymerase gene. By comparing the amount of HCMV DNA Polymerase PCR products in the MeDIP eluate compared with input DNA, we found that DNA from the AD169 strain was more abundant than the VR1814 strain, supporting the notion that AD169 is more methylated than VR1814 (results and methodological details in Fig. S2). In addition, we digested DNA from infected MRC-5 cells with HpaII or MspI, followed by a qRT-PCR using the same DNA Polymerase primers as for the MeDIP DNA. Since the sequence between the primers contains a CCGG site, the ratio between the qPCR products of the HpaII and MspI can be used as a measure for sequence-specific methylation. The results again showed that AD169 was more methylated in the tested sequence compared with the VR1814 strain (results and methodological details in Fig. S2). The apparent methylation assayed by LUMA, NNA and HCMV quantification ( and ) and the direct virus-specific methylation analysis (Fig. S2), suggest that newly replicated viral DNA from VR1814 infected cells avoids DNA methylation to a greater extent than does AD169.
Figure 3. Determination of HCMV genome relative copy numbers in infected cells by using real time qPCR. A specific region of the HCMV genome (pp150) was amplified by qPCR as reported previously.Citation29 18S genomic region was amplified as internal control for amount of host cell genome. HCMV copy numbers were normalized based on 18S amplification. Error bars denote standard deviation of triplicate measurements in triplicate infections, in total 9 data points for each viral strain.
HCMV infection alters intracellular localization of DNMTs in vitro and in vivo
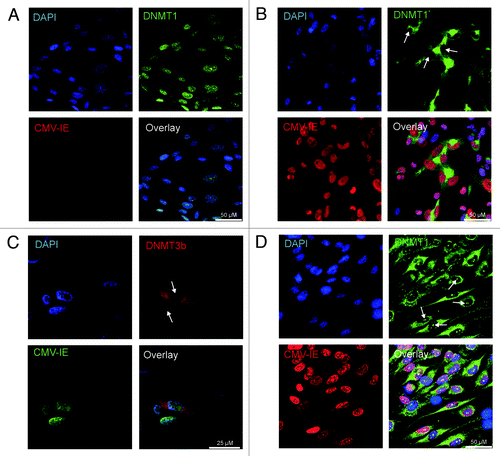
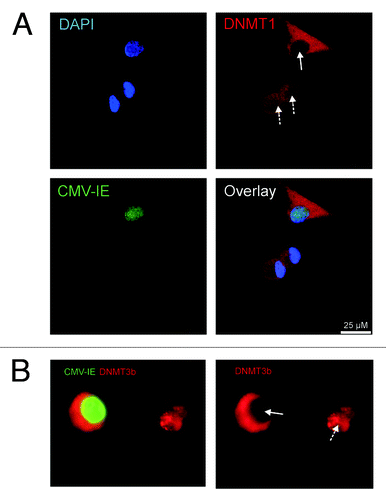
To further investigate the mechanism of HCMV induced hypomethylation, we analyzed the expression of DNMTs in HCMV infected cells by immunofluorescence staining. By using this method, a clear change in the intracellular localization of DNMT1 and 3b was evident after HCMV infection. While DNMT1 was localized in the nucleus of uninfected MRC-5 cells, HCMV-VR1814 infection caused the enzyme to shift and accumulate in the cytoplasm with DNMT1 virtually undetectable in the nucleus (). DNMT3b was undetectable in non-infected cells (data not shown), but also accumulated in the cytoplasm, similar to DNMT1, following HCMV infection (). Since the AD169 strain also displayed global hypomethylation effects by LUMA, albeit to a lesser extent, we assessed its intracellular localization by immunostaining, as well. Also this HCMV strain re-localized to the cytoplasm similarly to VR1814 (Fig. S3). As a control for the specificity of the nuclear exclusion of DNMT1, staining of the proliferating cell nuclear antigen PCNA was performed. As expected, PCNA was not shifted from the MRC-5 cell nuclei after HCMV infection, but it acquired a more patchy appearance in infected cells compared with the otherwise even nuclear staining pattern, likely reflecting viral DNA replication activity (Fig. S4).
Figure 4. Intracellular localization of DNMT1 and DNMT3b in fibroblasts infected by VR1814 at 3dpi. DNMT1 in uninfected cells (A), DNMT1 in infected cells (B), DNMT3b in infected cells (C), DNMT1 in Foscavir treated virus infected cells (D). In (A, B and D), HCMV-IE proteins were visualized by Texas red conjugated donkey anti mouse IgG (red) and DNMT1 with Alexa488 conjugated donkey anti rabbit IgG (green). In C, HCMV-IE proteins were visualized with Alexa488-conjugated donkey anti mouse IgG (green) and DNMT3b were visualized by Texas red conjugated donkey anti rabbit IgG (red). Representative images of cells with nuclear depletion and cytoplasmic localization of DNMT1 and DNMT3b are marked by arrows. Note that the size bar in A, B and D is 50 µm, while the size bar in C is 25 µm.
To examine whether viral early or late proteins controlled the re-localization of DNMT1, HCMV VR1814 infected MRC-5 cells were treated with Foscavir to inhibit expression of late viral genes, followed by assessment of DNMT1 by immunostaining. Foscavir treatment of virus-infected fibroblasts did not affect the majority of the virus-induced nuclear exclusion of DNMT1, which suggests that it is mainly dependent on viral IE gene products. However, a small and condensed staining pattern was visible in most infected cells treated with Foscavir (). This implies that a late viral gene product may also facilitate cytoplasmic localization of DNMT1, suggesting multiple viral strategies to control viral DNA methylation.
To address our findings in a more clinically relevant system, the localization of DNMTs was investigated in HCMV infected in vitro-differentiated macrophages. We also observed DNMT1 and DNMT3b re-localization to the cytoplasm following HCMV infection () in HCMV infected macrophages.
Figure 5. Intracellular localization of DNMT1 and DNMT3b in VR1814 infected macrophages. HCMV-IE proteins were visualized by Alexa488-conjugated donkey anti mouse IgG (green) and DNMT1 (A), and DNMT3b (B), were visualized by Texas red conjugated donkey anti rabbit IgG (red). For panel A, DAPI was used as counterstain of the nucleus (blue). Infected (plain arrow) and uninfected (dashed arrow) cells are shown.
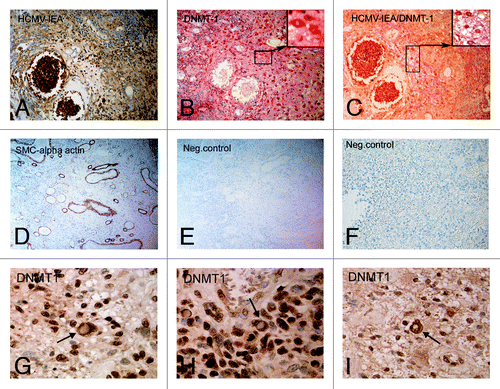
To further examine whether HCMV infection also affects the localization of DNMTs in vivo, we analyzed paraffin-embedded intestinal tissue sections from five HCMV positive patients with active inflammatory bowel disease. By using single and double immunostaining procedures we detected expression of DNMT1 in the cytosol of HCMV infected inflammatory cells in the bowel of patients while non-infected cells exhibited nuclear expression (). These data suggest that the altered intracellular DNMT1 localization is a consequence of HCMV infection also in clinical samples.
Figure 6. DNMT1 translocates from the nucleus in HCMV infected cells in intestinal tissue specimens obtained from IBD patients. Sections were stained for (A) HCMV-IE protein expression (brown, DAB, 20X magnification), (B) DNMT-1 protein expression (Red, Fast Red, 20X magnification), (C) HCMV/DNMT-1 double staining (Fast Red/DAB, 20X magnification), (D) SMC α-actin (DAB, 10X magnification), (E and F) Negative control without primary antibody. Cytoplasmic localization of DNMT1 was also investigated in 3 additional IBD patients. In all of them, cells with cytoplasmic localization of DNMT1 were observed as indicated by arrows (G-I).
Discussion
Epigenetic marks, including histone modifications and DNA methylation, are essential players in the gene regulating systems in eukaryotic cells.Citation30 An important strategy for viruses should therefore be to disrupt the epigenetic regulatory system of their host cells in favor of the viral functions. This would provide the opportunity for the virus to express viral genes and replicate their genome as well as to impair host cell protective mechanisms.Citation31 In this study, we examined whether the cellular DNA methylation machinery affects susceptibility to viral infection, and if HCMV infection affects DNA methylation. Our data demonstrate that: i) DNA methylation influences cellular susceptibility to HCMV infection; ii) human cytomegalovirus inhibits the host cell DNA methylation machinery and; iii) infection by human cytomegalovirus results in the re-localization of DNMTs from the nucleus to their accumulation in the cytoplasm.
A number of studies have reported that HCMV interacts with the host epigenetic systems. It has been shown that treatment of cells non-permissive to HCMV with histone deacetylase inhibitors converts these cells into permissive cells.Citation13 This may be related to the finding that the HCMV promoter/enhancer is associated with acetylated histone H4 during the lytic cycle of viral replication.Citation15,Citation32 In a murine model for MCMV infection, Hummel et al. also reported that DNA methylation inhibitor 5-Aza-2’-deoxycytosine treatment enhances induction of immediate early (IE) gene expression and reactivation of the virus.Citation16 Several reports also demonstrate a direct interaction of IE proteins with several chromatin modifying enzymes such as histone deacetylases HDAC1, HDAC2, HDAC3, and histone methyltransferases such as G9a and Suvar(3–9)H1.Citation33-Citation35
We addressed the inhibitory effect of DNMTs on HCMV infection by using the HCT-116 cell lines lacking DNMT1, DNMT3b or both. We observed that the HCMV-laboratory adapted strain AD169 was unable to infect HCT-116 cells while it could successfully infect the HCT-DKO cells, which implies a role for DNA-methylation in HCMV susceptibility (). In support of these findings, the resistant HCT-116 cells became susceptible to AD169 in a time dependent manner, after inhibition of DNA methylation by 5-azacytidine treatment (). This demonstrates that the lack of DNMT expression can be reproduced by reduced enzyme activity.
Since the clinical isolate VR1814 could infect the wild-type HCT-116 cells, it may possess some mechanism(s) to overcome the inhibitory effect of DNMTs. This observation sheds light over the unknown variable capacity of different HCMV strains to infect certain host cells. HCMV clinical isolates, such as VR1814, contain at least 19 genes that are not present in the laboratory strain AD169,Citation36 and may contain genes directly or indirectly responsible for overcoming host cell DNA methylation that are missing in AD169. Indeed, it was demonstrated by using qPCR that the copy number of the HCMV genomes was similar in AD169 and VR1814 infected MRC-5 cells (). Thus, the differences in DNA methylation observed by LUMA and NNA in cells infected with the two viral strains may be explained by the fact that newly replicated AD169 genomes become to some extent methylated in the host cells, in contrast to VR1814 genomes, which are apparently almost completely unmethylated. This issue was also interrogated by using virus-specific methylation analysis employing MeDIP as well as methylation sensitive restriction digestion, followed by quantification of HCMV DNA. We observed less abundance of methylated VR1814 DNA compared with AD169 DNA, supporting the notion that AD169 DNA is partially methylated while VR1814 is substantially less so.
Western blotting analysis of IE72 and IE86 protein expression after infection with the two strains showed that AD169 infected cells expressed mainly IE72, but none or small amounts of IE86, while VR1814 express both IE72 and IE86 at relatively equal amounts (Fig. S5). Whether the expression of IE86 is responsible for the ability of VR1814 to inhibit DNA methylation and/or play a role in making cells more susceptible to infection is currently under our further investigation.
We also show that the interference of the host cell methylation by HCMV infection is associated with a shift in intracellular localization from the nucleus to the cytoplasm, and a cytoplasmic accumulation of DNMT1 ( and ). DNMT3b was not detected in the control cells—likely because de novo methylation is not taking place in differentiated cells—but nevertheless accumulated in the cytoplasm upon infection. The draining of DNA methyltransferases from the nucleus may provide the opportunity for the virus to express viral proteins and replicate its genome in a DNA methylation free environment to avoid formation of suppressing complexes on viral promoters. Cytoplasmic localization of DNMT1 is not unique to HCMV infection. Previously, Cardoso and Leonhardt reported that DNMT1 is actively retained in the cytoplasm during early embryonic stage.Citation37 It is therefore possible that HCMV maintains DNMT1 outside the nucleus by taking advantage of a natural process of host cells, rather than directly interacting with DNMT1/DNMT3b. The appearance of cytoplasmic DNMT1 in infected macrophages as well as in intestinal epithelial cells of patients with inflammatory bowel disease demonstrated that accumulation of cytoplasmic DNMTs also occurs in infected cells in vivo.
We investigated if HCMV infection could affect genome-wide DNA methylation of the host cell by using the high throughput DNA methylation profiling system, Illumina Infinium bead chips (Illumina), which interrogates specific CpG methylation in approximately 14,000 gene promoters. The Illumina results demonstrated no significant methylation changes in any specific genomic locus in infected cells following infection (Fig. S1). The main reason for this may be that HCMV infection inhibits all the genomic replication activity in infected cells (and thus passive demethylation), and therefore the host cell is not undergoing any changes in specific loci. It still remains to be investigated, however, whether random DNA methylation alterations in individual cells occur due to the HCMV induced effects on the methylation machinery that cannot be detected in DNA collected from a cell population.
Several types of cancer are associated with viral infection, such as HCMV in glioblastoma, colon, breast and prostate cancer,Citation6,Citation7,Citation45,Citation48 HPV with cervical cancer, EBV with lymphoma and HBV with hepatocellular carcinoma.Citation31 Since HCMV is involved in both cancer and inflammatory diseases, such as IBD and cardiovascular disease, it is important to consider HCMV’s ability to manipulate the host cell epigenome, as part of specific pathological processes. In conclusion, we show that HCMV manipulates the host cell DNA methylation capacity by mechanisms that involve intracellular re-direction of DNMT1 and DNMT3b to the cytoplasm. Since inhibition of viral late protein expression only partially affected the re-localization process, we suggest that both early and late HCMV genes may be involved. Such complex mechanisms is supported by our finding that both tested viral strains resulted in cytoplasmic accumulation of DNMT1 despite that de novo virus methylation was different between the two. We propose that these findings may be important starting points for understanding mechanistic aspects of HCMV infection and epigenetic underpinnings of subsequent disease. Our findings may shed light on the epigenetic mechanisms that confer cells susceptible to HCMV infection/activation and may therefore point to devising new drugs for preventing both the previously established pathological effects on the immune system, and the novel malignant consequences of this common virus.
Materials and Methods
Cell culturing and viral infection
The colon carcinoma cell lines HCT-116 (wild-type), HCT-DNMT1KO (clone lacking DNMT1), HCT-3bKO (clone lacking DNMT3b), and HCT-DKO (clone lacking both DNMT1 and DNMT3b) were kindly provided by Dr. Bert Vogelstein (The Johns Hopkins Medical University, Baltimore, MD).Citation26 All cell lines were cultured in McCoy 5A medium supplied by 10% FBS and 100U penicillin and 100µg streptomycin (Gibco BRL, Sweden). At 50% confluency cells were infected in triplicates by either the HCMV laboratory strain AD169 (ATCC® VR-538, Manassas, VA USA) or the endothelial cell-adapted clinical isolate VR1814 strain (kindly provided by Professor Giuseppe Gerna, University of Pavia, Italy), at a multiplicity of infection (MOI) 1 and 10 for 24 h. Cells were fixed in acetone-methanol (1:1) and stained by mouse anti-immediate early monoclonal antibody and DAPI to evaluate infection levels by fluorescence microscopy. Pictures were taken and the number of infected cells of 1,000 counted cells was calculated. The experiments were performed three times.
For treatment of HCT-116 cells by 5-azacytidine (Sigma), the cells were cultured in 8 well chamber slides. At 50% confluency, 5-azacytidine was added at final concentration of 10 µM for 24, 48 and 72 h or left untreated. Cells were then infected with the HCMV-laboratory adapted strain AD169 for 24 h, and then fixed and stained for HCMV-IE proteins as mentioned above.
The human embryonic lung fibroblast cell line MRC-5 (ATCC, CCL-171) was maintained in DMEM with glutamine, supplemented with 100U penicillin and 100µg streptomycin per ml (Gibco BRL, Grand Island, NY) and used during passages 22–26. At 80% confluence, cells were challenged by either HCMV-AD169, or HCMV-VR1814 at MOI of 5, or were left un-infected. After 3 d of infection at 37°C and 5% CO2, cells were harvested and analyzed as indicated. Foscavir® treatment was done by adding 5 mM Foscavir (AstraZeneca, Sweden) 2 h before infection to inhibit the expression of late viral gene products.Citation39
Peripheral blood mononuclear cells (PBMCs) from healthy blood donors were isolated by density gradient centrifugation on Lymphoprep™ (Axis-Shield PoC AS, Oslo, Norway). Lymphocytes were washed in sterile Hanks’ Balanced Salt Solution (Sigma-Aldrich Gmbh, Steinheim, Germany) twice, and were reconstituted in IMDM medium (GIBCO), supplemented with L-Glutamine, 25 mM HEPES buffer, 1% penicillin/streptomycin (Gibco) and 10% heat inactivated human AB-serum. Cells were plated in 8-well chamber slides (LAB-TEKII, Nalgene-Nunc, Naperville, IL, USA) and incubated for 2 h. Non-adherent cells were removed; remaining cells were stimulated during the next 24 h for macrophage differentiation by adding a supernatant containing cytokines produced by allogenic T-cells and monocytes, so called “allo-cytokines” as described previously.Citation40 After 24h, cells were washed repeatedly in 1x phosphate buffered saline (PBS), followed by the addition of “60/30/10” medium (60% of Serum Free Lymphocyte Medium AIM V® (Gibco), 30% of IMDM medium with the addition of L-Glutamine, 25 mM HEPES, 1% PEST and 10% human AB-serum) to the cells, and incubated for 7 d at 37°C and 5% CO2. Differentiated macrophages were then infected at MOI of 10 for 96 h, with the VR1814 strain, or were left un-infected. After incubation, cells were washed in 1 x PBS, and fresh “60/30/10” medium was added to the cells and cultured for an additional 3 d.
DNA isolation
Genomic/viral DNA was isolated using the Sigma Mammalian Genomic DNA Isolation Kit. DNA concentration was determined by a Nanodrop ND-100 instrument.
Luminometric Methylation Assay (LUMA)
Global methylation analysis of CCGG sequences was performed using LUMA. Genomic DNA was extracted using GenElute® mammalian genomic DNA miniprep kit (Sigma, USA). LUMA assay was performed as described earlier.Citation27,Citation41 Briefly, 200–500 ng genomic DNA was digested by HpaII + EcoRI or MspI + EcoRI restriction endonucleases for 4 h at 37°C. The extent of DNA digestion was determined by polymerase extension assay using a 96MA Pyrosequencing™ platform (Biotage, Sweden). HpaII/EcoRI and MspI/EcoRI ratios were calculated by the corresponding A and G+C peaks and DNA methylation level was defined as HpaII/MspI. Genomic Methylation percentage was calculated as 100(1-HpaII/MspI).Citation27
5-Methylcytosine quantification by Nearest Neighbor Analysis
The total level of CpG methylation was quantified by nearest neighbor analysis (NNA) using α32P-dGTP as described previously.Citation42 The intensity of 5-methylcytosine, and cytosine mononucleotide spots on thin layer chromatography was measured using a phosphoimager (Fujifilm, Japan) and the Image Gauge™ analysis software (Fujifilm, Japan). Percentage of DNA methylation levels were defined as 100x[methyl-cytosine]/ [methyl-cytosine + cytosine].
Immunofluorescence and confocal microscopy
Cells cultured in eight-well chamber slides were fixed with ice cold methanol:acetone (1:1) for 10 min, washed in 1xPBS and then air-dried. After blocking with 1% bovine serum albumin (USB, Cleveland, OH) in 1x PBS for 20 min, slides were incubated for 1 h at room temperature with a monoclonal antibody specific for HCMV immediate early proteins (Argene Parc Technologique, Delta Sud, France, 1/150 dilution), and polyclonal rabbit antibodies specific for DNMT1 (ab19905, Abcam, Cambridge, UK, 1/200 dilution). Polyclonal rabbit antibodies specific for DNMT3b (ab2851) and for PCNA (ab2426) were from Abcam and were both used in 1/200 dilution. Following 3 washes in PBS for 5 min, AlexaFluor 488-conjugated donkey anti-mouse IgG (1/500 dilution, Molecular probe) and Texas Red-conjugated donkey anti-rabbit antibody (1/500 dilution, Molecular probe) was added and incubated at room temperature for 60 min. Slides were washed 3 times for 5 min with 1xPBS. Cell nuclei were stained with DAPI (Sigma, USA), prior to mounting the slides with fluorescent anti fade mounting medium (DakoCytomation, Glostrup, Denmark). Negative control slides were processed in the same manner, but omitting the primary antibodies. Also a DNMT1 blocking peptide (ab21999, Abcam, Cambridge, UK) was used to assess specificity of the DNMT1 antibodies. Slides were analyzed and photographed by fluorescence microscopy using a Leica TCS SP5 confocal laser-scanning microscope with a 40x objective (Leica, Germany). For Alexa488, Ar laser at 488nm was used for excitation, and the detection range was 494–540nm. For Texas red, a HeNe laser at 594nm was used for excitation, and the detection range was 596–650 nm. For nuclear visualization by DAPI, a 405 nm diode laser was used for excitation, and detection range was 420–460 nm. The pinhole was set to 67.9 μM.
Immunohistochemistry
Paraffin embedded intestinal tissues sections from five HCMV-infected patients with active ulcerative colitis were analyzed for expression of HCMV proteins and DNMT-1 by using immunohistochemistry technique as described previously.Citation5 Primary antibodies used were: mouse anti-HCMV-IE (Chemicon), rabbit anti–human DNMT1 (ab19905 AbCam), mouse anti–human SMC -actin (DakoCytomation, Glostrup, Denmark). For detection of HCMV-IE and SMC -actin proteins, colorimetric determination was performed with a three-step horseradish peroxidase detection system (BioGenex) with the chromogen diaminobenzidine (DAB) (Innovex Sciences). DNMT1 was detected using a streptavidin alkaline phosphatase detection system (DakoCytomation) and visualized by FastRed (DakoCytomation) in double staining, and DAB for single staining. Hematoxylin (Sigma-Aldrich) was used for counterstaining and slides were mounted in permanent mounting medium (DakoCytomation). Slides were analyzed by a Leica DMRB Light microscope equipped with a DC-480 CCD camera for image capture.
Additional material
Download Zip (1.5 MB)Acknowledgments
This study was financially supported by the Torsten Söderbergs Foundation, The Swedish Heart and Lung Foundation, the Swedish Cancer Foundation, the Karolinska Institutet, and the Stockholm County Council. We thank Dr. Koon Yaiw for helpful discsussions and experimental advices. M. Zeitelhofer was supported by the Wenner-Gren Foundation.
Ethical statement
The ethical committees at Huddinge Hospital (diary no. 38/95) and Karolinska Institutet (EPN2006/764–32) approved the studies on the subject of inflammatory bowel disease.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Sinclair J, Sissons P. Latency and reactivation of human cytomegalovirus. J Gen Virol 2006; 87:1763 - 79; http://dx.doi.org/10.1099/vir.0.81891-0; PMID: 16760381
- Söderberg-Naucler C. Human cytomegalovirus persists in its host and attacks and avoids elimination by the immune system. Crit Rev Immunol 2006; 26:231 - 64; PMID: 16928188
- Corrado E, Novo S. Role of inflammation and infection in vascular disease. Acta Chir Belg 2005; 105:567 - 79; PMID: 16438065
- Kandiel A, Lashner B. Cytomegalovirus colitis complicating inflammatory bowel disease. Am J Gastroenterol 2006; 101:2857 - 65; http://dx.doi.org/10.1111/j.1572-0241.2006.00869.x; PMID: 17026558
- Cobbs CS, Harkins L, Samanta M, Gillespie GY, Bharara S, King PH, et al. Human cytomegalovirus infection and expression in human malignant glioma. Cancer Res 2002; 62:3347 - 50; PMID: 12067971
- Harkins L, Volk AL, Samanta M, Mikolaenko I, Britt WJ, Bland KI, et al. Specific localisation of human cytomegalovirus nucleic acids and proteins in human colorectal cancer. Lancet 2002; 360:1557 - 63; http://dx.doi.org/10.1016/S0140-6736(02)11524-8; PMID: 12443594
- Samanta M, Harkins L, Klemm K, Britt WJ, Cobbs CS. High prevalence of human cytomegalovirus in prostatic intraepithelial neoplasia and prostatic carcinoma. J Urol 2003; 170:998 - 1002; http://dx.doi.org/10.1097/01.ju.0000080263.46164.97; PMID: 12913758
- Hollink N, Dzabic M, Wolmer N, Boström L, Rahbar A. High prevalence of an active human cytomegalovirus infection in patients with colonic diverticulitis. J Clin Virol 2007; 40:116 - 9; http://dx.doi.org/10.1016/j.jcv.2007.07.008; PMID: 17765008
- Cinatl J Jr., Cinatl J, Vogel JU, Rabenau H, Kornhuber B, Doerr HW. Modulatory effects of human cytomegalovirus infection on malignant properties of cancer cells. Intervirology 1996; 39:259 - 69; PMID: 9078467
- Strååt K, Liu C, Rahbar A, Zhu Q, Liu L, Wolmer-Solberg N, et al. Activation of telomerase by human cytomegalovirus. J Natl Cancer Inst 2009; 101:488 - 97; http://dx.doi.org/10.1093/jnci/djp031; PMID: 19318640
- Michaelis M, Doerr HW, Cinatl J. The story of human cytomegalovirus and cancer: increasing evidence and open questions. Neoplasia 2009; 11:1 - 9; PMID: 19107226
- Dunn W, Chou C, Li H, Hai R, Patterson D, Stolc V, et al. Functional profiling of a human cytomegalovirus genome. Proc Natl Acad Sci U S A 2003; 100:14223 - 8; http://dx.doi.org/10.1073/pnas.2334032100; PMID: 14623981
- Murphy JC, Fischle W, Verdin E, Sinclair JH. Control of cytomegalovirus lytic gene expression by histone acetylation. EMBO J 2002; 21:1112 - 20; http://dx.doi.org/10.1093/emboj/21.5.1112; PMID: 11867539
- Nitzsche A, Paulus C, Nevels M. Temporal dynamics of cytomegalovirus chromatin assembly in productively infected human cells. J Virol 2008; 82:11167 - 80; http://dx.doi.org/10.1128/JVI.01218-08; PMID: 18786996
- Reeves MB, Lehner PJ, Sissons JG, Sinclair JH. An in vitro model for the regulation of human cytomegalovirus latency and reactivation in dendritic cells by chromatin remodelling. J Gen Virol 2005; 86:2949 - 54; http://dx.doi.org/10.1099/vir.0.81161-0; PMID: 16227215
- Hummel M, Yan S, Li Z, Varghese TK, Abecassis M. Transcriptional reactivation of murine cytomegalovirus ie gene expression by 5-aza-2′-deoxycytidine and trichostatin A in latently infected cells despite lack of methylation of the major immediate-early promoter. J Gen Virol 2007; 88:1097 - 102; http://dx.doi.org/10.1099/vir.0.82696-0; PMID: 17374752
- Ioudinkova E, Arcangeletti MC, Rynditch A, De Conto F, Motta F, Covan S, et al. Control of human cytomegalovirus gene expression by differential histone modifications during lytic and latent infection of a monocytic cell line. Gene 2006; 384:120 - 8; http://dx.doi.org/10.1016/j.gene.2006.07.021; PMID: 16989963
- Kauder SE, Bosque A, Lindqvist A, Planelles V, Verdin E. Epigenetic regulation of HIV-1 latency by cytosine methylation. PLoS Pathog 2009; 5:e1000495; http://dx.doi.org/10.1371/journal.ppat.1000495; PMID: 19557157
- Goldberg AD, Allis CD, Bernstein E. Epigenetics: a landscape takes shape. Cell 2007; 128:635 - 8; http://dx.doi.org/10.1016/j.cell.2007.02.006; PMID: 17320500
- Miranda TB, Jones PA. DNA methylation: the nuts and bolts of repression. J Cell Physiol 2007; 213:384 - 90; http://dx.doi.org/10.1002/jcp.21224; PMID: 17708532
- Feinberg AP, Tycko B. The history of cancer epigenetics. Nat Rev Cancer 2004; 4:143 - 53; http://dx.doi.org/10.1038/nrc1279; PMID: 14732866
- Strickland FM, Richardson BC. Epigenetics in human autoimmunity. Epigenetics in autoimmunity - DNA methylation in systemic lupus erythematosus and beyond. Autoimmunity 2008; 41:278 - 86; http://dx.doi.org/10.1080/08916930802024616; PMID: 18432408
- Stenvinkel P, Karimi M, Johansson S, Axelsson J, Suliman M, Lindholm B, et al. Impact of inflammation on epigenetic DNA methylation - a novel risk factor for cardiovascular disease?. J Intern Med 2007; 261:488 - 99; http://dx.doi.org/10.1111/j.1365-2796.2007.01777.x; PMID: 17444888
- Yi-Deng J, Tao S, Hui-Ping Z, Jian-Tuan X, Jun C, Gui-Zhong L, et al. Folate and ApoE DNA methylation induced by homocysteine in human monocytes. DNA Cell Biol 2007; 26:737 - 44; http://dx.doi.org/10.1089/dna.2007.0619; PMID: 17764386
- Shi H, Wang MX, Caldwell CW. CpG islands: their potential as biomarkers for cancer. Expert Rev Mol Diagn 2007; 7:519 - 31; http://dx.doi.org/10.1586/14737159.7.5.519; PMID: 17892361
- Rhee I, Bachman KE, Park BH, Jair KW, Yen RW, Schuebel KE, et al. DNMT1 and DNMT3b cooperate to silence genes in human cancer cells. Nature 2002; 416:552 - 6; http://dx.doi.org/10.1038/416552a; PMID: 11932749
- Karimi M, Johansson S, Stach D, Corcoran M, Grandér D, Schalling M, et al. LUMA (LUminometric Methylation Assay)--a high throughput method to the analysis of genomic DNA methylation. Exp Cell Res 2006; 312:1989 - 95; http://dx.doi.org/10.1016/j.yexcr.2006.03.006; PMID: 16624287
- Jones PA. DNA methylation and cancer. Cancer Res 1986; 46:461 - 6; PMID: 2416425
- Strååt K, de Klark R, Gredmark-Russ S, Eriksson P, Söderberg-Nauclér C. Infection with human cytomegalovirus alters the MMP-9/TIMP-1 balance in human macrophages. J Virol 2009; 83:830 - 5; http://dx.doi.org/10.1128/JVI.01363-08; PMID: 18945772
- Jones PA, Baylin SB. The epigenomics of cancer. Cell 2007; 128:683 - 92; http://dx.doi.org/10.1016/j.cell.2007.01.029; PMID: 17320506
- Li HP, Leu YW, Chang YS. Epigenetic changes in virus-associated human cancers. Cell Res 2005; 15:262 - 71; http://dx.doi.org/10.1038/sj.cr.7290295; PMID: 15857581
- Reeves MB, MacAry PA, Lehner PJ, Sissons JG, Sinclair JH. Latency, chromatin remodeling, and reactivation of human cytomegalovirus in the dendritic cells of healthy carriers. Proc Natl Acad Sci U S A 2005; 102:4140 - 5; http://dx.doi.org/10.1073/pnas.0408994102; PMID: 15738399
- Nevels M, Paulus C, Shenk T. Human cytomegalovirus immediate-early 1 protein facilitates viral replication by antagonizing histone deacetylation. Proc Natl Acad Sci U S A 2004; 101:17234 - 9; http://dx.doi.org/10.1073/pnas.0407933101; PMID: 15572445
- Reeves M, Murphy J, Greaves R, Fairley J, Brehm A, Sinclair J. Autorepression of the human cytomegalovirus major immediate-early promoter/enhancer at late times of infection is mediated by the recruitment of chromatin remodeling enzymes by IE86. J Virol 2006; 80:9998 - 10009; http://dx.doi.org/10.1128/JVI.01297-06; PMID: 17005678
- Park JJ, Kim YE, Pham HT, Kim ET, Chung YH, Ahn JH. Functional interaction of the human cytomegalovirus IE2 protein with histone deacetylase 2 in infected human fibroblasts. J Gen Virol 2007; 88:3214 - 23; http://dx.doi.org/10.1099/vir.0.83171-0; PMID: 18024889
- Cha TA, Tom E, Kemble GW, Duke GM, Mocarski ES, Spaete RR. Human cytomegalovirus clinical isolates carry at least 19 genes not found in laboratory strains. J Virol 1996; 70:78 - 83; PMID: 8523595
- Cardoso MC, Leonhardt H. DNA methyltransferase is actively retained in the cytoplasm during early development. J Cell Biol 1999; 147:25 - 32; http://dx.doi.org/10.1083/jcb.147.1.25; PMID: 10508852
- Cobbs CS, Soroceanu L, Denham S, Zhang W, Britt WJ, Pieper R, et al. Human cytomegalovirus induces cellular tyrosine kinase signaling and promotes glioma cell invasiveness. J Neurooncol 2007; 85:271 - 80; http://dx.doi.org/10.1007/s11060-007-9423-2; PMID: 17589804
- Wahren B, Oberg B. Inhibition of cytomegalovirus late antigens by phosphonoformate. Intervirology 1980; 12:335 - 9; http://dx.doi.org/10.1159/000149093; PMID: 6244238
- Söderberg-Nauclér C, Streblow DN, Fish KN, Allan-Yorke J, Smith PP, Nelson JA. Reactivation of latent human cytomegalovirus in CD14(+) monocytes is differentiation dependent. J Virol 2001; 75:7543 - 54; http://dx.doi.org/10.1128/JVI.75.16.7543-7554.2001; PMID: 11462026
- Karimi M, Johansson S, Ekström TJ. Using LUMA: a Luminometric-based assay for global DNA-methylation. Epigenetics 2006; 1:45 - 8; http://dx.doi.org/10.4161/epi.1.1.2587; PMID: 17998810
- Ramsahoye BH. Nearest-neighbor analysis. Methods Mol Biol 2002; 200:9 - 15; PMID: 11951658
- Pierer M, Rothe K, Quandt D, Schulz A, Rossol M, Scholz R, et al.. Anti-cytomegalovirus seropositivity in rheumatoid arthritis is associated with more severe joint destruction and more frequent joint surgery. Arthritis Rheum 2011; In press; http://dx.doi.org/10.1002/art.34346; PMID: 22183424
- Pérez-Mercado AE, Vilá-Pérez S. Cytomegalovirus as a trigger for systemic lupus erythematosus. J Clin Rheumatol 2010; 16:335 - 7; PMID: 20859222
- Harkins LE, Matlaf LA, Soroceanu L, Klemm K, Britt WJ, Wang W, et al.. Detection of human cytomegalovirus in normal and neoplastic breast epithelium. Herpesviridae 2010; 1:8; PMID: 21429243
- Nitzsche A, Paulus C, Nevels M. Temporal dynamics of cytomegalovirus chromatin assembly in productively infected human cells. J Virol 2008; 82:11167 - 80; PMID: 18786996
- Ernberg I, Karimi M, Ekström TJ. Epigenetic mechanisms as targets and companions of viral assaults. Ann N Y Acad Sci 2011; 1230:E29 - 36; PMID: 22417105
- Rahbar A, Stragliotto G, Orrego A, Peredo I, Taher C, Willems J, et al.. Low levels of Human Cytomegalovirus Infection in Glioblastoma Multiforme associates with patient survival; -a case-control study. Herpesviridae 2012; 16:3; PMID: 22424569