Abstract
In plants, heterochromatin is maintained by a small RNA-based gene silencing mechanism known as RNA-directed DNA methylation (RdDM). RdDM requires the non-redundant functions of two plant-specific DNA-dependent RNA polymerases (RNAP), RNAP IV and RNAP V. RNAP IV plays a major role in siRNA biogenesis, while RNAP V may recruit DNA methylation machinery to target endogenous loci for silencing. Although small RNA-generating regions that are dependent on both RNAP IV and RNAP V have been identified previously, the genomic loci targeted by RNAP V for siRNA accumulation and silencing have not been described extensively. To characterize the RNAP V-dependent, heterochromatic siRNA-generating regions in the Arabidopsis genome, we deeply sequenced the small RNA populations of wild-type and RNAP V null mutant (nrpe1) plants. Our results showed that RNAP V-dependent siRNA-generating loci are associated predominately with short repetitive sequences in intergenic regions. Suppression of small RNA production from short repetitive sequences was also prominent in RdDM mutants including dms4, drd1, dms3 and rdm1, reflecting the known association of these RdDM effectors with RNAP V. The genomic regions targeted by RNAP V were small, with an estimated average length of 238 bp. Our results suggest that RNAP V affects siRNA production from genomic loci with features dissimilar to known RNAP IV-dependent loci. RNAP V, along with RNAP IV and DRM1/2, may target and silence a set of small, intergenic transposable elements located in dispersed genomic regions for silencing. Silencing at these loci may be actively reinforced by RdDM.
Background
Heterochromatin, highly condensed chromosomal DNA associated with repetitive sequences and transposons, appears to play important roles in nuclear processes such as chromosomal segregation and genomic stability.Citation1-Citation3 Studies from fission yeast and plants have demonstrated a role for small RNA in heterochromatin formation and maintenance.Citation4-Citation7 In the plant silencing mechanism called RNA-directed DNA methylation (RdDM), siRNA directs de novo cytosine methylation at homologous DNA regions.Citation8,Citation9 These RNA-based silencing mechanisms provide an important level of epigenetic control to repress transposable elements (TEs) and aberrant genes such as transgenes, while also playing a role to regulate the expression of endogenous genes.
In plants, two nuclear DNA-dependent RNA polymerases have evolved to work exclusively in RNA-mediated silencing pathways.Citation10 DNA-dependent RNA polymerases IV and V (RNAP IV and RNAP V) each have a unique largest subunit (NRPD1 and NRPE1, respectively) yet share the second largest subunit (NRPD2/NRPE2) with each other;Citation11 they also have many subunits shared or paralogous to those in RNAP II.Citation12 While mutations of NRPD1, NRPE, and NRPD2/NRPE2 lack an obvious phenotypic impact, selective activation of transposons and other repeats in mutants has been observed.Citation13-Citation16 RNAP IV functions upstream of RdDM along with RNA-DEPENDENT RNA POLYMERASE 2 (RDR2) and DICER-LIKE 3 (DCL3) to generate 24 nt siRNA from heterochromatic loci, while RNAP V functions downstream with ARGONAUTE 4 (AGO4)-associated siRNA to facilitate de novo DNA methylation at siRNA target loci.Citation16-Citation20 Interestingly, recent work has demonstrated that RNAP II transcriptional activity is also involved in siRNA-directed gene silencing via interactions with an AGO4-siRNA complex.Citation21,Citation22 Although it is unclear how RNAP lV, RNAP V and RNAP II activities are functionally integrated in heterochromatin silencing, these studies suggest a delicate coordination and yet functional diversification of these three polymerases in RdDM.
RdDM likely has three major steps: (1) siRNA biogenesis, (2) production of non-coding transcripts as a scaffold and (3) assembly of AGO-siRNA effector complexes to recruit methylation machinery and target genomic loci.Citation23 In siRNA biogenesis, heterochromatic regions are likely transcribed by RNAP IV, made double-stranded by RDR2, processed into 24 nt siRNA by DCL3, and finally the siRNA population is incorporated by AGO4 and probably AGO6 to form an AGO-siRNA complex.Citation24-Citation26 At some of these loci, RNAP V likely generates non-coding transcripts that could serve as the scaffolds.Citation21 Other RdDM effectors involved in scaffold RNA generation include RNAP II, an IWR1-like transcriptional factor DEFECTIVE IN MERISTEM SILENCING 4 (DMS4),Citation27 and the “DDR complex,” which contains the SNF2-like chromatin-remodeling factor DEFECTIVE IN RNA-DIRECTED-DNA-METHYLATION 1 (DRD1), a structural-maintenance-of-chromosomes hinge domain-containing protein DEFECTIVE IN MERISTEM SILENCING 3 (DMS3), and the novel protein RNA-DIRECTED DNA METHYLATION 1 (RDM1), which binds a methylated single-stranded DNA.Citation15,Citation28-Citation30 The resulting scaffold RNAs plus the AGO-siRNA complex form a guiding complex together with an SPT5-like transcriptional elongation factor (KOW-DOMAIN CONTAINING TRANSCRIPTION FACTOR 1, KTF1), the zinc-finger domain protein INVOLVED IN DE NOVO 2 (IDN2), and the DDR complex.Citation27,Citation31-Citation33 Finally, through a mechanism that is not fully understood, the guiding complex recruits DNA methyltransferases and histone methyltransferases, such as DOMAINS REARRANGED METHYLASE 1 and 2 (DRM1 and DRM2), CHROMOMETHYLASE 3 (CMT3), SUPRESSOR OF VARIEGATION 3–9 HOMOLOG 9 (SUVH9) and SUVH2 to direct the silencing of specific genomic loci,Citation34-Citation37 which results in both the generation and maintenance of heterochromatin.
Unlike RNAP IV, it is less clear how RNAP V functions in siRNA production. It has been suggested that the role of RNAP V in RNA-directed gene silencing is to promote DNA methylation by recruiting the silencing complex to siRNA-targeted loci.Citation15,Citation16,Citation19,Citation28 Yet, studies have shown that RNAP V mutants have reduced small RNAs at some loci, indicating a non-redundant role of RNAP V to RNAP IV in siRNA accumulation.Citation16,Citation19,Citation21 Heterochromatic silencing may require two rounds of siRNA production; while RNAP IV is necessary for the production of primary siRNA for de novo methylation at endogenous loci, RNAP V targeting is crucial for the production of siRNA from methylated loci for reinforcement and possibly spreading of methylation.Citation9,Citation16,Citation38 Although siRNA production seems to largely rely on RNAP IV action, the presence of RNAP IV-dependent but RNAP V-independent siRNA-generating loci implies the existence of different RNAP IV- and RNAP V-directed siRNA activity at endogenous loci.Citation16,Citation19,Citation39 Analysis of transgene-targeting siRNAs shows that a loss of function of RdDM proteins, including DMS4, DRD1, DMS3 and RDM1, which are responsible for scaffold RNA generation, impacts secondary siRNA production from the region downstream of the RdDM target, 24 nt siRNA biogenesis from RNAP IV- and RNAP V-dependent loci, and recruitment of RdDM machinery by RNAP V.Citation22,Citation28,Citation40 Thus, we hypothesized that RNAP V may direct siRNA generation and target methylation at specific genomic loci, possibly representing endogenous equivalents to the transgene-derived secondary siRNAs.
To characterize RNAP V-dependent, heterochromatic siRNA-generating regions in Arabidopsis, we employed next-generation sequencing to deeply sequence the small RNA population in wild-type and RNAP V null mutant (nrpe1) plants. Our results showed that NRPE1-dependent siRNA-generating loci are associated predominately with short repetitive sequences in intergenic regions, with an overrepresentation of short interspersed elements (SINEs) and rolling circle/helitron (RC/Helitron) sequences. Suppression of small RNA production from short repetitive sequences was also prominent in RdDM mutants, including dms4, drd1, dms3 and rdm1 mutants, indicating an equivalency with RNAP V. The genomic regions targeted by RNAP V were generally quite small, with the average impacted region spanning only a few hundred base pairs. Our results suggest that RNAP V affects siRNA production from specific loci with genome features dissimilar to known RNAP IV-dependent loci, indicating that RNAP V operates on specific genomic loci for siRNA production in RdDM.
Results
Identification of > 2,000 NRPE1-dependent small RNA loci by small RNA profiling
To investigate the role of RNAP V in RdDM, we analyzed small RNA from Arabidopsis immature inflorescences of wild-type Columbia (ecotype Col-0, hereafter “Col”) and a mutant of the largest RNAP V subunit, NRPE1 (allele nrpb1b-11, hereafter “nrpe”). We generated small RNA libraries from three biological replicates of Col or nrpe samples (Table S1) for Illumina sequencing. The raw small RNA sequences were trimmed to remove adaptor sequences and matched to the Arabidopsis genome (TAIR version 9). In order to compare between libraries, the abundance of each small RNA was normalized to reads per five million (RP5M). For replicates of Col and nrpe libraries, we obtained 1.3 to 6.2 million total genome-matched small RNA sequences, corresponding to 0.5 to 2.1 million ‘distinct’ sequences that are different sequences, found uniquely in the data set. We used a Spearman’s correlation to assess the reproducibility of small RNA data sets in replicates by pairwise comparison (Fig. S1). The correlation coefficient rho among Col or nrpe libraries was high (approximately 0.73 to 0.80), indicating a strong correlation among replicate libraries. The proportion of distinct vs. total genome-matched reads represented the degree of divergence and complexity of the small RNA population being captured in the library, which was from 0.346 to 0.455 for wild-type and from 0.350 to 0.439 for the nrpe samples (Table S1A). The small RNA complexity is much lower compared with the results of a previous study by Mosher et al.Citation19 (Table S1B), probably resulting from the ~100-fold increase of our sequencing depth due to improved technologies. More importantly, the proportion of distinct small RNA reads was similar between wild-type and nrpe mutant libraries (median values 0.346 and 0.352, respectively), in agreement with the results from Mosher et al. (Table S1). The reduction in the small RNA complexity in nrpe libraries was minimal compared with prior reports of substantial reductions in an Arabidopsis rdr2 mutant (proportion of distinct = 0.04 or 0.05) (Table S1).Citation41 In wild-type Arabidopsis inflorescences, the proportion of 24 to 21 nt small RNA abundances is typically about 3:1, in which most 24-mers are a diverse population of heterochromatic siRNAs and most 21-mers are highly abundant miRNAs.Citation39 Our results showed that the proportion of 24 nt small RNA abundances were reduced in all nrpe libraries compared with all wild-type libraries () and to a ratio of 1.62:1 for the averaged replicates (), a reduction of more than 1.5 fold in the nrpe data set. This impact in 24 nt siRNA is more reminiscent of an rdr2 mutant, but at a much lower degree of severity, as the Arabidopsis rdr2 mutant showed a ratio of 0.16:1 for 24 to 21 nt abundances which was a nearly complete reduction (18-fold) over wild-type.Citation41
Figure 1. Small RNA-generating loci are suppressed in nrpe mutants. (A) Small RNA size profiles in wild-type (Col) and a null mutant of NRPE1 (nrpe) replicate libraries, normalized to the percentage of 21 nt abundance in wild-type libraries. For each size class, small RNA abundance (excluding structural RNA) was calculated as a percentage to the sum of abundances of total genome-matched reads. (B) Averaged percentage of abundances in small RNA size classes, calculated from data in (A); “Col avg” or “nrpe avg” indicate the averaged values for each set of three libraries. (C) Number of clusters impacted in the nrpe mutant, based on summed small RNA abundances per cluster. For each cluster, we compared the average of small RNA HNA of three Col libraries to the average of small RNA HNA of three nrpe libraries. The ratio of Col vs. nrpe is used when the HNA of Col libraries is greater than that in nrpe libraries, while the ratio of nrpe vs. Col is used when the HNA of nrpe libraries is greater than that in Col libraries (shown as negative values). The inset graph (note the reduced y-axis) expands to the full range of the fold differences to explain the high value of the “≥ 10” column; the basis for the high “≥10” bar is a very long tail of low frequency clusters highly impacted in nrpe. (D) Genic vs. non-genic and repeat- vs. non-repeat-associated characteristic of RNAP V-dependent small RNA clusters based on the TAIR version 9 annotations. RNAP V-dependent (“RNAP V-dpt”) and RNAP V-independent (“RNAP V-indpt”) clusters were defined as described in text. A total of 11,667 small RNA-generating, 2,201 RNAP V-dependent and 7,680 RNAP V-independent clusters were analyzed.
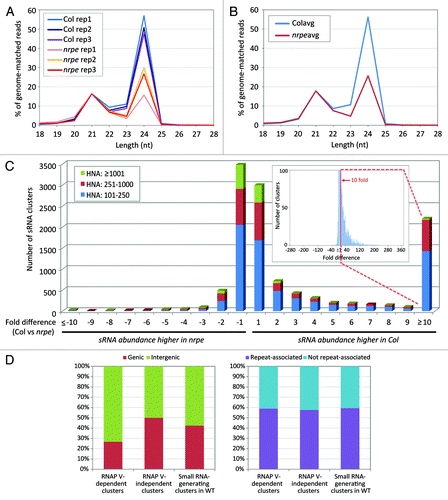
Given the incomplete reduction in 24 nt siRNA abundances and a minimal impact on complexity, yet knowing that RNAP V has a known but secondary role in heterochromatic siRNA biogenesis, we were curious to know which genomic loci showed reduced siRNA levels in the nrpe mutant. To identify the RNAP V-dependent, siRNA-generating regions in the genome, we deployed a proximity-based algorithm to group and quantify clusters of small RNAs. In Arabidopsis, a total of 239,339 adjacent, non-overlapping fixed-size (500 bp) bins or “clusters” were defined to cover the complete nuclear genome. A value of “hits-normalized-abundance” (HNA) was calculated by dividing the normalized abundance (in RP5M) for each small RNA by the number of genomic locations to which the small RNA maps (its “hits”). In the cluster analysis, the HNA values for all the small RNA that mapped to a given 500 bp cluster was summed separately for each library regardless of the small RNA sizes. Finally, the sum of abundance in each cluster was averaged for the three Col replicate libraries and compared with the averaged value of three nrpe libraries. Since we had demonstrated a strong correlation among the three replicates, we used the set of averaged replicate library abundances for all subsequent analyses. The cluster analysis summarizes the small RNA abundance within specific genomic regions where siRNA may be produced, and also provides a practical way to compare the same genomic region across different libraries. Furthermore, each cluster was annotated with gene and repeat information (e.g., TAIR9 annotated retrotransposons and DNA transposons), which allowed us to characterize the genomic features associated with particular small RNA-generating loci.
Based on the result of the cluster analysis, we identified genomic loci for which small RNA production was dependent on RNAP V. We selected 11,667 clusters from a total of 239,339 clusters at which the sum of HNA of Col and nrpe libraries was greater than a baseline value (HNA = 100 RP5M), excluding the regions from which minimal small RNAs are produced (Table S2). Next, we compared the small RNA abundance of each individual cluster in wild-type and nrpe libraries and calculated the fold difference of HNA between them. As shown in and Table S2, the majority of the small RNA-generating loci were not impacted in the nrpe mutant, with 65% of the clusters showing a less than 2-fold difference to the control. Only 25 small RNA-generating clusters were greatly upregulated in nrpe compared with the wild-type, with at least 10-fold higher levels in nrpe (a review of these loci demonstrated no obvious pattern or significant characteristic, and thus these were not considered further). Notably, we observed a clear preference toward downregulated clusters in the nrpe mutant, with 18% of the total (2,201 clusters) exhibiting at least a 10-fold higher small RNA abundance in the Col compared with nrpe libraries (). An examination of a randomly-selected set of clusters with small RNA abundances at least 10-fold higher in wild-type demonstrated quite substantial differences (50-fold or more) can exist between the mutant and wild-type (Table S3). Next, all small RNA-producing clusters were classified based on overall abundances in wild-type and nrpe libraries ( and Table S2). In the nrpe-suppressed loci, the abundance of small RNAs per impacted cluster was lower than those not impacted in the mutant; 1,427 out of 2,201 (65%) had a summed HNA from 101 to 250 RP5M, and 745 out of 2,201 (34%) had a summed HNA from 250 to 1000 RP5M. Similarly, only 29 of 1,171 clusters with RP5M (2.5%) exceeding 1,000 RP5M were greatly impacted in nrpe (Table S2). Thus among the 2,201 clusters impacted in nrpe, the effect was greater on small RNA clusters of low to moderate abundance. For the purposes of this analysis, we defined these 2,201 clusters as ‘RNAP V-dependent,’ since their small RNA accumulation was reduced at least 10-fold or more in the absence of NRPE1.
Overrepresentation of SINE and RC/Helitron repeats in RNAP V-dependent small RNA loci
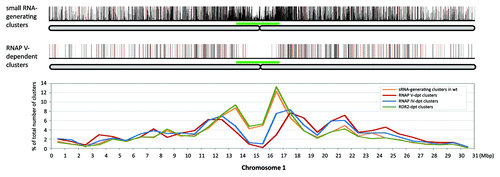
We were interested to characterize the type of genomic regions dependent on RNAP V for small RNA biogenesis, which represented only 18% of all of the small RNA-generating loci in the Arabidopsis genome. In addition, we defined 7,680 clusters, which showed minimal changes between the Col and nrpe data sets (+/− 2-fold difference in HNA) as ‘RNAP V-independent’ clusters. First, the RNAP V-dependent and RNAP V-independent clusters were categorized based on the gene and repeat annotations. For small RNA-generating clusters in the wild-type libraries, less than half (42%) were located in genic regions and 58% of the clusters were in intergenic regions, suggesting a slight intergenic disposition (). For the RNAP V-independent clusters, half were located in genic regions while the other half were in intergenic regions. However, only 27% of RNAP V-dependent clusters were in genic regions while the majority (73%) was in intergenic regions, indicating a strong intergenic pattern. When we compared the clusters to repeat annotations, RNAP V-dependent clusters were not more repetitive than RNAP V-independent clusters or than small RNA-generating clusters in wild-type (). This result was not unexpected, as previous studies had focused on intergenic regions as the targets of RNAP V and RNAP II activity in RdDM.Citation21,Citation28 Prior reports had implied that RNAP V-dependent regions might be more distal to the centromeres,Citation19 so we examined the genomic distribution of these RNAP V-dependent clusters. In wild-type Arabidopsis, small RNAs are known to be most abundant in the transposable element-rich, pericentromeric regions of the chromosomes (and S2, upper and lower panels).Citation42,Citation43 The distribution of RDR2-dependent clusters from an analysis of an rdr2 mutantCitation43 coincided with the distribution of small RNA-generating clusters in wild-type plants, with both exhibiting a strong pericentromeric localization. On the other hand, RNAP V-dependent clusters had less pericentromeric concentration (and S2, middle and lower panels) in comparison to the RDR2-dependent clusters, suggesting a more dispersed and possibly euchromatic chromosomal distribution of the RNAP V-dependent loci generating these small RNAs. The genomic locations of RNAP V-dependent and RNAP IV-dependent clusters were very similar (and S2); while the Mosher et al. (2008) analysis suggested a pericentromeric bias for RNAP IV-dependent region, our data are not directly comparable due to the low depth of their sequencing data. Nonetheless, both our and their studies indicate that RNAP V-dependent loci are predominantly dispersed across the chromosomes in non-pericentromeric regions.
Figure 2. Genome-wide distributions of RNAP V-dependent small RNA-generating loci. Upper panel, the distribution of small RNA-generating loci and small RNA abundance in wild-type along chromosome 1 with bar height indicating the sum of HNA in Col libraries; red bars indicate HNA greater than 500; full-height bars are HNA between 100 to 500, and shorter bars are HNA < 100. The chromosome is illustrated below in gray, with approximate pericentromeric regions based on centromeric staining data marked with green bars.Citation51,Citation62 Middle panel, differentially-expressed RNAP V- dependent small RNA-generating clusters; full-height bars in black are clusters with reduction relative to Col of between 10- to 50-fold; red bars indicate clusters with fold reduction > 50. Lower panel, the density of RNAP V-dependent clusters across chromosome 1, plotted as the percentage of number of clusters in 1 Mb windows over the total number of RNAP V-dependent clusters mapped to chromosome 1. The distribution of small RNA-generating clusters and NRPD1(RNAP IV)- and RDR2-dependent clusters were plotted for comparison. Data for chromosomes 2 to 5 are shown in Figure S2.
Since heterochromatic siRNAs produced by RNAP IV and RNAP V were shown to be associated with certain repetitive sequences,Citation16,Citation19 we examined the types of repeats impacted in nrpe. We analyzed the 2,201 RNAP V-dependent clusters and 7,680 RNAP V-independent clusters using the repeat annotation in Arabidopsis TAIR9 genome,Citation44 which allowed us to associate individual clusters with any overlapping repeat type. We should note that we also performed this analysis using repeats identified by RepeatMasker,Citation45 and our conclusions were not substantially different (data not shown); the analyses described here were generated using the TAIR-annotated repeats. Among the clusters generating small RNAs at a level greater than 100 RP5M, 59% of clusters were repeat-associated while 41% (4,740 out of 11,667) were not associated with known repeats (Table S4). Similarly, 59% of RNAP V-dependent clusters were repeat-associated while 41% (904 out of 2,201) of these clusters were not associated with repetitive sequences. The percentage of repeat-associated clusters was quite similar (42% to 41%) between RNAP V-independent and dependent clusters (Table S4), suggesting that RNAP V-dependent, siRNA-generating regions were no more or less repetitive compared with either wild-type or RNAP V-independent regions.
Although the overall level of repeat association is similar between RNAP V-dependent and independent loci, certain classes of repeats were much more predominant in RNAP V-dependent clusters than the control set and vice versa. Most notably, SINEs were associated with 0.1% of RNAP V-independent clusters, but they were 7.1% of RNAP V-dependent clusters, a 54-fold difference (). The RC/Helitron, DNA/Mariner, DNA/hAT, DNA/Harbinger, and LINE/L1 elements were all represented at a higher percentage in RNAP V-dependent loci than in RNAP V-independent loci ( and Table S4). On the other hand, LTR/Copia- and LTR/Gypsy-associated clusters were underrepresented in RNAP V-dependent clusters, with the greatest difference of representation as 16.1% in the control set vs. 1.0% in RNAP V-dependent clusters for LTR/Gypsy repeats (a 17-fold difference). Therefore, fewer LTR-associated clusters were represented in highly suppressed small RNA-generating loci in the RNAP V mutants, indicating that small RNA production from LTR-associated loci was less likely to depend on RNAP V.
Figure 3. SINEs are overrepresented in nrpe-suppressed siRNA-generating loci. (A) Classes of repetitive sequences represented in RNAP V-dependent small RNA-generating clusters. RNAP V-dependent and RNAP V-independent clusters are defined as described in the legend. Repetitive sequences are identified based on Arabidopsis TAIR9 repeat annotation. Percentage of repeat-annotated clusters to the numbers of RNAP V-dependent or RNAP V-independent loci is shown. Change of repeat class representation is calculated by the ratio of its percentage in RNAP V-dependent vs. RNAP V-independent cluster (in red) or by the ratio of its percentage in RNAP V-independent vs. RNAP V-dependent cluster (in blue), which indicates whether the repeat class is over-represented or under-represented in RNAP V-dependent loci, respectively. (B) Overrepresentation of SINEs in RNAP V-dependent clusters. Number of SINE-annotated clusters is plotted against the fold difference in small RNA abundance between wild-type and nrpe libraries. Number of RC/Helitron-annotated clusters, another repeat class that is overrepresented in RNAP V-dependent clusters, is also plotted.
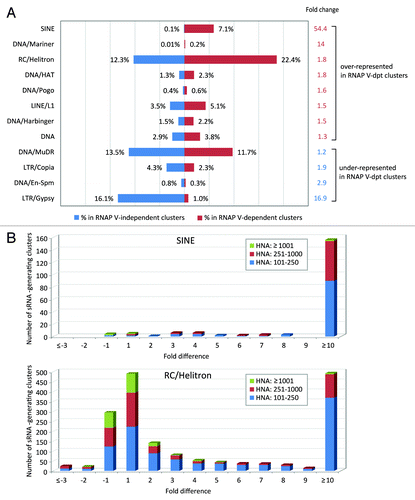
Our results showed that SINE was the most overrepresented repeat class associated with RNAP V-dependent clusters compared with the RNAP V-independent clusters. RC/Helitron, the most abundant repeat class associated with RNAP V-dependent clusters, are also overrepresented by nearly 2-fold in RNAP V-dependent loci. SINE and RC/Helitron elements are highly abundant, small-sized transposable elements in the Arabidopsis genome, which led us to ask how many of these overlap small RNA-generating clusters affected by the nrpe mutation. Most of the 188 small RNA-generating, SINE-associated clusters had a low-to-medium abundance, and 156 (83%) were greatly impacted in nrpe mutants with at least a 10-fold reduction in the small RNA abundances ( and Table S4). Similarly, 28% (492 out of 1757) of small RNA-generating RC/Helitron-type loci showed greatly reduced small RNA abundances in nrpe mutant ( and Table S4). Although 11% of RC/Helitron-type small RNA-generating loci had high levels of small RNA abundances (HNA ≥ 1001 RP5M), almost all the RC/Helitron-associated loci impacted in the RNAP V mutant had a low to moderate level of small RNAs, suggesting that the RNAP V dependency was greater for SINE- or RC/Helitron-type clusters of low-to-moderate small RNA abundance (). For other overrepresented repeats in nrpe, such as DNA/Mariner and DNA/hAT, a substantial proportion of their small RNA-generating loci were also strongly suppressed in nrpe (57% for DNA/Mariner and 25% for DNA/hAT) (Table S4). Conversely, only 10% to 1.6% of small RNA-generating clusters of LTR-type repeats (LTR/Copia and LTR/Gypsy) were impacted in RNAP V mutants. Taken together, these results showed that repeat classes including SINEs exhibit high RNAP V dependency since 20% to 80% of their small RNA-generating loci were heavily suppressed in the nrpe mutant. We found it curious that different classes of transposable elements (e.g., DNA transposons and retrotransposons) were affected in nrpe mutant in a similar fashion. The relatively small size of some of these types of elements suggested a possible correlation between the repeat length and the impact on small RNA abundance.
RNAP V-dependent small RNA-generating loci comprise short genomic regions
The enrichment of short repeat classes such as SINE and RC/Helitron and the dearth of long LTR species in RNAP V-dependent loci led us to speculate about the role of repeat size in RNAP V dependency. Prior reports have indicated that it may be difficult to maintain the silenced state of regions of just a few nucleosomes in length;Citation46 hence, we suspected that RNAP V may be responsible for the silencing of small repetitive regions. We used several approaches to analyze the relationship between size and RNAP V-dependency. In the first such analysis, we conducted a comparison between the total length of the repeat vs. the reduction in the small RNA abundances in the nrpe mutant. The repeat element boundaries (start and end coordinates) were defined by the repeat annotation in TAIR9 genome, and the total hits-normalized small RNA abundance was determined for each repeat element for both wild-type and nrpe libraries. We selected 3,826 elements that had a sum of small RNA abundances above the baseline (HNA of Col > 100 and HNA of nrpe > 5 RP5M). For these elements, we plotted the fold difference of HNA between wild-type and mutant libraries vs. the lengths of the repeat elements (). The result showed that the majority of highly nrpe-suppressed small RNA-generating loci were associated with repetitive sequences 5 kb or less in length. We next plotted the fold difference of HNA from repeat elements under 5 kb in 500 bp increments in length (). About one-third (1,395 out of 3,826) of repeat elements shorter than 1 kb produced small RNAs and, from those, 536 repeat elements showed at least a 10-fold reduction of small RNA abundance in nrpe mutant (). These data indicate that the length of the repeats was inversely correlated with the degree of nrpe-suppression in small RNA abundances, and small RNA production was mostly impacted in the nrpe mutant at shorter repeats.
Figure 4. Repeat length is reversely correlated with the suppression of small RNA production in the nrpe mutant. (A) Plot of nrpe-dependent suppression of small RNA abundance vs. the length of repetitive sequences. The HNA was summed for the abundance of small RNAs mapped to the length of each annotated repetitive sequence, i.e., small RNA abundance for each repeat. Y-axis shows the Log10 scale of fold difference of sum of HNA between wild-type and nrpe libraries. The best-fit trendline from the power regression is showed. The section within 5 kb repeat length is further expanded and shown in (B). (B) Degree of nrpe-dependent suppression. The width of boxes is proportional to the square root of the numbers of repeats. (C) Numbers of RNAP V-dependent, small RNA-generating repeats (dark red) vs. the total number of small RNA-generating repeats (light red) in different repeat classes are shown along with the percentage (the former vs. the latter). (D) Comparison of numbers of RNAP V-dependent, small RNA-generating repeats (red) and the total number of small RNA-generating repeats (light red) across the size of repeat in selected repeat families. (E) Length profiles of repeat-annotated small RNA-generating elements are plotted with fixed-width box. White boxes represent data from small RNA-generating elements, and red boxes represent data from RNAP V-dependent, small RNA-generating elements based on the criteria described in the main text. Median values for both data sets are shown. Repeat length between 250 bp to 300 bp is highlighted in blue. For box plots in (B) and (E), the box indicates the fold changes in the 25th to 75th percentile with the center bar indicating the median. The dashed lines indicate the range of values in the lower or upper quartiles, terminating at the minimum or maximum lengths.
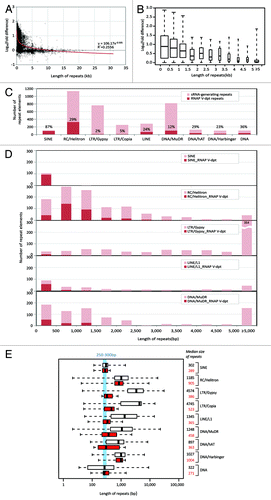
Next, we analyzed the repeat length within transposon families relative to the impact in the nrpe mutant. Transposons are remarkably heterogeneous in size, copy number and diversity within a repeat family.Citation47,Citation48 To investigate whether the negative correlation between repeat size and RNAP V dependency also occurred within transposon families, we first separated the 3,826 elements into different repeat classes and plotted the number of nrpe-suppressed elements vs. the total number of small RNA-generating repeat elements (). RC/Helitron and SINE had the highest numbers of repeats suppressed in the nrpe mutant; the impact of nrpe affected more than 87% of small RNA-generating SINEs (). This result agreed with the trend of nrpe-suppression we observed in the static cluster analysis using a fixed width of 500 bp (Table S4). Once the transposable elements were categorized into repeat classes, we examined nrpe-directed suppression using small RNA-generating repeats grouped by a repeat length in a 500 bp increment (and S3). For every repeat class we examined in the nrpe mutant, it was clear that small RNA production was mostly impacted from shorter members of the repeat class. Therefore, our data indicated that the RNAP V dependency may be mostly determined by the size of the repeats regardless of the repeat types.
To assess the correlation of repeat length and nrpe-directed siRNA suppression, we calculated the median length of repeats generating small RNAs in wild-type and the subset of elements greatly impacted in nrpe for each repeat class (). The range of sizes in the genome for each repeat class varies: LTR/Gypsy and LTR/Copia are the longest with a median exceeding 2 kb; RC/Helitron, LINE/L1 and several types of DNA transposons have median around 1 kb; and the median size of SINEs is around 300 bp (, white boxes). When the size ranges of RNAP V-dependent elements were plotted in comparison, it was clear that RNAP V-dependent clusters were predominantly the subset of shorter elements within each repeat type (, red boxes), which agreed with our previous results (). Perhaps the most revealing one was the LTR/Gypsy class: although these repeat elements are the least impacted in the nrpe mutant (), the 22 LTR/Gypsy elements demonstrating RNAP V-dependent small RNA suppression of 1,322 small RNA-producing LTR/Gypsy elements are substantially smaller in size (). A similar trend was observed for all the larger elements including LTR/Copia, LINE/L1 and DNA/MuDR. It is worth noting that there was little difference in impacted SINE sizes relative to all SINEs, perhaps because the SINEs as a class have a median size of just 302 bp. In summary, our data showed that the nrpe mutation affects small RNA accumulation for smaller repeat elements across all repeat classes ().
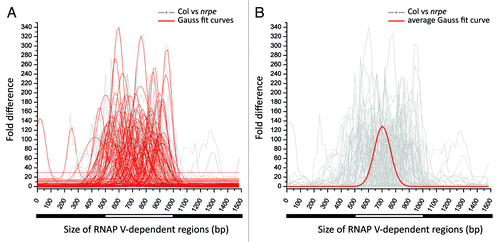
We sought to determine the approximate width of loci reduced in small RNAs in the nrpe mutant as a surrogate for a measurement of RNAP V transcript length, as such transcripts have not yet been identified on a genomic scale. From the 2,201 clusters defined above, we selected 100 RNAP V-dependent clusters that had highly differential small RNA levels in Col vs. nrpe (HNA of Col > 250 and HNA of nrpe = 0 to 5 RP5M). Consistent with our observation that RNAP V-dependent regions are predominantly small, almost all of these clusters are distal to each other in the genome. For each of these 100 clusters, we extracted its 500 bp genomic sequence and flanking 500 bp to obtain a 1500 bp region in total, and we plotted the fold-difference between Col and nrpe small RNA abundances for each of the 100 clusters using a sliding window of 100 bp across the region (gray lines in ). Next, the distribution of the fold difference for each of the RNAP V-dependent region was fitted into smooth peaks using Gaussian distribution fit; a single curve representing the average small RNA abundance difference and the width of the region of impacted small RNAs was fitted to the data at each cluster (, red lines in the foreground). The fitted curves were positioned mainly in the 500 bp central region with slight spread 100 bp upstream or downstream from the center, indicating that the nrpe-directed suppression is concentrated within each 500 bp regions we selected. Using the height and width of each fitted curve, an average curve was calculated () to represent the average width and RNAP V-dependent small RNA abundance for these 100 clusters. The average width of a RNAP V-dependent locus was ~238 bp (), the size of a typical RNAP V-dependent locus in the genome, possibly representing the length of a region in which RNAP V is active.
Figure 5. Assessment of the size of RNAP V-dependent regions. (A) Plot showing the degree of nrpe-dependent suppression on small RNA abundance across the width of RNAP V- dependent regions. One hundred of 500 bp, highly RNAP V-dependent clusters are selected (the average sum of HNA in nrpe libraries is less than 5 and HNA in Col is greater than 250 RP5M). Each selected cluster is joined together with its 500 bp upstream and 500bp downstream region for a 1,500 bp region. The degree of nrpe-dependent small RNA suppression is presented as the rounded fold-difference between Col vs. nrpe libraries in a window of 100 bp, sliding every 20 bp across the entire 1,500 bp region (gray background lines). For each of the RNAP V-dependent region, Gaussian distribution is used to fit the gray line of fold difference and shown as the red curve in the foreground. (B) The average curve was calculated (in red) from the data in (A) to represent the average size of a RNAP V-dependent region. The average width of a RNAP V-dependent locus was computed from the individual height and widths of each fitted curve in (A), independent of the exact position within the cluster (positions were ignored to focus on the shape of the curve for each cluster). The width of the average curve was calculated as 238 bp.
SINE and RC/Helitron overrepresentation in other RdDM mutants
RdDM effectors such as DMS4, DRD1, DMS3 and RDM1 play a role in scaffold RNA production and de novo methylation and maintenance, possibly by promoting RNAP V transcription and activation. Because of the functional relationship of these proteins in RdDM, we were interested in the small RNA profiles of mutants of these genes. These mutants have previously been examined only in selected genomic loci.Citation22,Citation26,Citation27 We generated small RNA libraries from mutant alleles dms4–1, drd1–1, dms3–1 and rdm1–4, which have well-characterized epigenetic phenotypes, such as reduced 24 nt siRNA and cytosine methylation.Citation22,Citation27,Citation30,Citation40 We also generated another control library [“Wt (T + S)” for wild-type plus the target and silencer transgene construct] for these RdDM mutants, which were described in a previous mutant screen that assayed the release of transgene silencing.Citation40 From the deep sequencing results, the abundance of each small RNA was normalized to reads per five million (RP5M) (Table S1). We obtained 5 to 16 million total genome-matched small RNA sequences, corresponding to 1.3 to 2.7 million distinct sequences. The small RNA complexity of the RdDM mutant libraries ranged from 0.151 to 0.269, which were at similar levels to the 0.264 of the Wt (T+S) control library. The size distribution profiles of the RdDM mutant compared with the control showed the reduction of 24 nt abundances in drd1–1, dms3–1 and rdm1–4 mutant libraries but not in the dms4–1 library (, and Table S5). Interestingly, dms4–1 showed a slightly lower ratio of 24:21 nt abundance to the control library, indicating that the reduction of 24 nt siRNA abundance was minimal in the absence of DMS4 (). However, the reduction in the percentage of 24 nt abundance was more than 3-fold greater in drd1–1, dms3–1 and rdm1–4 libraries compared with their control, indicating a significant impact on heterochromatic siRNA accumulation in these three components of the DDR complex as in the nrpe mutant. The greater impact of drd1–1, dms3–1 and rdm1–4 on 24 nt abundance may be explained by the fact that two to four times more of the high abundant small RNA sequences were greatly reduced in drd1–1, dms3–1 and rdm1–4 libraries than in the dms4–1 library, especially for 24 nt small RNAs (Table S5A). As a result, there is a greater percentage of the high-abundance 24 nt small RNA impacted in drd1–1, dms3–1 and rdm1–4 libraries than in the dms4–1 library (Table S5B).
Figure 6. RdDM mutants show similar trends to the nrpe mutant of suppression of SINE-annotated siRNA-generating clusters. (A) Reduction of 24 nt small RNAs in RdDM mutant libraries. Shown for control [“Wt (T+S)”] and RdDM mutant libraries (drd1–1, drm3–1, rdm1–4 and dms4–1) are the 24 nt to 21 nt ratios of genome-matched small RNAs abundance (excluding structural RNAs). (B) Small RNA size profiles in control and RdDM mutant libraries. For each size class of small RNAs, the percentage of the small RNA abundance (excluding structural RNA) to the abundance of total genome-matched reads was calculated and normalized to the abundance of the 21 nt of wild-type libraries. (C) Number of clusters impacted in the RdDM mutants by the fold differences of small RNA abundance between control and RdDM mutant libraries. For each cluster, the fold difference of hit-normalized small RNA abundance between the control and RdDM libraries are calculated and rounded. Total numbers of clusters were tallied by the fold differences and plotted as data in Y-axis for each RdDM mutant library. These data and in particular the category of ≥ 10 is analogous to . (D) Pairwise comparison of RNAP V and RdDM effector-dependent. Numbers of clusters were calculated based on different criteria in nrpe, drd1–1, dms3–1, rdm1–4, dms4–1 libraries. Number of small RNA-generating clusters (criteria A) and RdDM effector-dependent, small RNA-generating clusters (criteria B) are shown for each RdDM mutant library. Pairwise comparison of nrpe vs. RdDM libraries (criteria C) shows the number of clusters which small RNA abundance is both greatly reduced in nrpe and RdDM mutant such as drd1- 1, i.e., RNAP V- and DRD1-dependent small RNA-generating clusters. (E and F) Area-proportional Venn diagrams show the number of small RNA-generating clusters which are suppressed in nrpe, dms4–1, and dms3–1 (E) and drd1–1, dms3–1, and rdm1–4 (F, left). The number of each sector in the Venn diagram in (f) is also shown in a representative Venn diagram (F, right). (G) Classes of repetitive sequences represented in RdDM effector-dependent small RNA-generating clusters. RNAP V-dependent (RNAP V-dpt) and RNAP V-independent (RNAP V-indpt) clusters are defined as described in the legend. RdDM effector-dependent clusters were selected with criteria B as described above. Repetitive sequences were identified by the TAIR9 repeat annotation. Proportion of repeat-annotated clusters to total number of RdDM-dependent loci for each control-RdDM mutant pair is shown on the y-axis (percentage).
![Figure 6. RdDM mutants show similar trends to the nrpe mutant of suppression of SINE-annotated siRNA-generating clusters. (A) Reduction of 24 nt small RNAs in RdDM mutant libraries. Shown for control [“Wt (T+S)”] and RdDM mutant libraries (drd1–1, drm3–1, rdm1–4 and dms4–1) are the 24 nt to 21 nt ratios of genome-matched small RNAs abundance (excluding structural RNAs). (B) Small RNA size profiles in control and RdDM mutant libraries. For each size class of small RNAs, the percentage of the small RNA abundance (excluding structural RNA) to the abundance of total genome-matched reads was calculated and normalized to the abundance of the 21 nt of wild-type libraries. (C) Number of clusters impacted in the RdDM mutants by the fold differences of small RNA abundance between control and RdDM mutant libraries. For each cluster, the fold difference of hit-normalized small RNA abundance between the control and RdDM libraries are calculated and rounded. Total numbers of clusters were tallied by the fold differences and plotted as data in Y-axis for each RdDM mutant library. These data and in particular the category of ≥ 10 is analogous to Figure 1C. (D) Pairwise comparison of RNAP V and RdDM effector-dependent. Numbers of clusters were calculated based on different criteria in nrpe, drd1–1, dms3–1, rdm1–4, dms4–1 libraries. Number of small RNA-generating clusters (criteria A) and RdDM effector-dependent, small RNA-generating clusters (criteria B) are shown for each RdDM mutant library. Pairwise comparison of nrpe vs. RdDM libraries (criteria C) shows the number of clusters which small RNA abundance is both greatly reduced in nrpe and RdDM mutant such as drd1- 1, i.e., RNAP V- and DRD1-dependent small RNA-generating clusters. (E and F) Area-proportional Venn diagrams show the number of small RNA-generating clusters which are suppressed in nrpe, dms4–1, and dms3–1 (E) and drd1–1, dms3–1, and rdm1–4 (F, left). The number of each sector in the Venn diagram in (f) is also shown in a representative Venn diagram (F, right). (G) Classes of repetitive sequences represented in RdDM effector-dependent small RNA-generating clusters. RNAP V-dependent (RNAP V-dpt) and RNAP V-independent (RNAP V-indpt) clusters are defined as described in the Figure 1 legend. RdDM effector-dependent clusters were selected with criteria B as described above. Repetitive sequences were identified by the TAIR9 repeat annotation. Proportion of repeat-annotated clusters to total number of RdDM-dependent loci for each control-RdDM mutant pair is shown on the y-axis (percentage).](/cms/asset/1cde4c60-4fba-4e57-8b6e-ed18affe03de/kepi_a_10920290_f0006.gif)
Next, we characterized the small RNA-generating regions that were dependent on the set of four RdDM effectors, DMS4, DRD1, DMS3, and RDM1. We first calculated the number of small RNA-generating clusters in four individual RdDM mutant libraries (sum of HNA control + mutant greater than 100 RP5M) (, criteria A), which identified approximately 14,000 to 18,000 clusters compared with the 11,667 small RNA-generating clusters in nrpe mutants. Next, we calculated the fold difference of HNA of small RNA clusters between control and each of four RdDM mutants, and plotted the numbers of clusters against the fold difference to see the impact of each mutation on small RNA abundance. As we observed for the nrpe libraries, the majority of the small RNA-generating loci were not impacted in the four RdDM mutants, with more than 50% of the clusters showing a less than 2-fold difference relative to the control (). On the other hand, a significant proportion of clusters were downregulated in the four RdDM mutants, with 8% to 16% of the total showing at least a 10-fold higher small RNA abundances in the control compared with the four RdDM mutant libraries (, criteria B). This result suggested that, although there were more small RNA generating loci in the four RdDM mutant libraries, the number of mutant-dependent loci was smaller in the four RdDM mutants than in nrpe, i.e., the degree of mutant-dependent suppression on small RNA abundance was less severe in the dms4–1, drd1–1, dms3–1 and rdm1–4 mutants.
We were interested to determine whether these small RNA loci suppressed in the four RdDM mutants were also suppressed in the nrpe mutant, which would indicate the degree of overlapping dependency of RNAP V and other RdDM effectors on small RNA production. Therefore, pairwise comparisons were done for small RNA clusters that were both highly suppressed in nrpe and in one of the four RdDM mutants (, criteria C). For the drd1–1 mutant, 1,067 out of 1,871 (57%) drd1-suppressed clusters were also suppressed in nrpe. Overall, we found around 60% of dms3-, rdm1- or dms4-suppressed clusters were also suppressed in nrpe, a high degree of overlap. There were 860 small RNA-generating clusters that were suppressed in nrpe, dms4–1 and dms3–1 (), and 1,348 clusters suppressed in drd1–1, dms3–1 and rdm1–4 (). Therefore, our data showed that dms3, rdm1 and drd1 were impacted predominantly at the same small RNA-generating regions, and these regions were in fact a subset of dms4-suppressed regions. This is consistent with their function together in the DDR complex.Citation49
Finally, we categorized these RdDM mutant-suppressed small RNA clusters according to their association with genomic repeats. We observed a similar trend of repeat types suppressed in these four RdDM mutant libraries as in nrpe: SINEs and RC/Helitrons were overrepresented in RNAP V-dependent and RdDM effector-dependent loci (). Conversely, LTR/Copia- and LTR/Gypsy-associated clusters were underrepresented in RNAP V-dependent and RdDM effector-dependent clusters (). Therefore, our results showed that small RNA production was impacted in shorter repeats not only in nrpe mutant but also in RdDM mutants including dms4, drd1, dms3 and rdm1.
Discussion
We characterized the RNAP V-dependent, small RNA-generating loci in Arabidopsis by deep sequencing of small RNAs in wild-type and a null RNAP V mutant allele nrpe. The loss-of-function mutation in the NRPE1 gene caused a substantial reduction in 24 nt small RNA accumulation in about 18% of small RNA-generating loci. RNAP V seems to affect small RNA production mainly from intergenic regions in euchromatin: 73% of RNAP V-dependent loci are in intergenic regions and 27% are in gene coding regions. We also examined the genome distribution of RNAP V-dependent loci. Pericentromeric regions are dense regions of repetitive sequences and thus heavily methylated and rich in heterochromatin.Citation50,Citation51 We show that RNAP V- and RNAP IV-dependent loci both exhibited a less-pericentromeric concentration especially in comparison to RDR2-dependent loci, and RNAP V-and RNAP IV-dependent loci appeared to be more dispersed across the genome rather than enriched at heterochromatin-dense regions near centromeres. The dispersed distribution of both RNAP V- and RNAP IV-dependent loci is reminiscent of the DRM1/2 target regions along the chromosome, which is in contrast to the CMT3- and KYP-target loci, which reside mostly in the centromere-proximal 2 Mb.Citation52 While non-CG methylation is generally maintained by CMT3 in pericentric heterochromatin, non-CG methylation is maintained by DRM1/2 DNA methyltransferases particularly in small, silenced regions that span only few nucleosomes.Citation6,Citation34 Collectively, our results suggested that RNAP V, along with RNAP IV and DRM1/2, may work via RdDM to target a subset of small transposable elements located in intergenic regions dispersed across the genome for silencing, which are likely maintained by non-CG methylation.
Apart from their similarity in chromosomal position, and as-yet uncharacterized features such as epigenetic marks, the differences between RNAP V- and RNAP IV-dependent loci apparently include the repeat type. Both RNAP IV- and RNAP V-dependent loci were shown to be associated with transposable repeat sequences, but the repeat associations differed between the two classes of loci ().Citation16,Citation19 LTR retrotransposons were overrepresented in RNAP IV-dependent and RNAP V-independent loci, while non-LTR retrotransposons and helitrons were overrepresented in RNAP IV-and RNAP V-dependent loci.Citation16,Citation19 Our results showed an overrepresentation of SINEs and RC/Helitrons and the underrepresentation of the gypsy class of retroelements at RNAP V-dependent loci. One plausible explanation for the different repeat types associated with RNAP IV or RNAP V may come from our results showing an inverse correlation between repeat size and nrpe repression. RNAP IV-directed silencing is clearly important for the repression of long repetitive sequences such as LTR elements.Citation16,Citation19 On the other hand, RNAP V-directed silencing appears to be focused on small repeats such as SINEs and RC/Helitrons (). As proposed previously by Zilberman and Henikoff,Citation46 shorter silenced regions of few nucleosomes in length might be difficult to maintain during replication due to the small number of epigenetic marks on a short piece of DNA and its associated histones, therefore requiring active siRNA targeting to reinforce silencing. In this case, RNAP IV may be required for siRNA production and RNA-directed silencing to suppress larger repetitive elements, whereas our data support the point that RNAP V may be essential for siRNA production in smaller and/or more euchromatic regions. Ahmed et al.Citation53 showed that, among the repeat classes, more than 90% of gypsy elements are densely methylated, SINEs are moderately methylated (75% methylated and 15% unmethylated), and RC/Helitrons are the least methylated sequences (40% methylated and 40% unmethylated). Their results were in agreement with the special patterns of repeat association of RNAP V-dependent small RNA-generating clusters that we observed. In other words, LTRs could be subjected to robust silencing by RNAP IV to render them heavily methylated in heterochromatic regions, while RC/Helitrons and SINEs, targeted by RNAP V, may direct silencing at TEs found more frequently in euchromatin.
This concept is reminiscent of a model of the reversible silencing of euchromatic genes by RNAP V action.Citation54 Indeed, Huettel et al.Citation54 proposed that RNAP V, along with DRD1, establishes a basal methylation state of the targets such as euchromatic promoters or intergenic repeats in gene-rich regions, which can be reversed when methylation marks are changed in response to developmental cues or environmental stresses. On the other hand, sequences such as LTRs in repeat-rich regions are subjected to additional epigenetic modification and persistent suppression in order to maintain the stable silencing state of heterochromatin. Taken together, these results suggest that RNAP V-directed, siRNA-mediated silencing may provide a dynamic regulation of DNA methylation by targeting repeats and/or enhancers in promoters or intergenic regions, which in turn regulate the expression of neighboring genes.
Our results indicated that RNAP V-dependent regions are smaller repeat-associated regions in Arabidopsis genome with an average size of ~238 bp. The fact that nrpe mutation greatly impacted smaller repeat-associated siRNA-generating regions across all repeat classes implies a relevance of repeat size in RNAP V-targeted silencing. Size has been a factor in the study of the relationship between nucleosome positioning and DNA methylation; nucleosome positioning strongly affects the patterns of DNA methylation throughout the genome and the 147 bp periodicity of methylation patterns matches the length of DNA wrapped around one nucleosome.Citation55 The estimated length of RNAP V-dependent regions could represent the length of more than one nucleosome, but more data are needed to link the size of RNAP V-dependent loci to nucleosome positioning and methylation periodicity. One related insight about the small size of RNAP V-dependent repeat-associated regions may come from the study of siRNA and methylated TEs.Citation53 Ahmed et al.Citation53 showed that unmethylated and poorly methylated TE sequences are smaller than their methylated counterparts, with the former having a medium size of less than 500 bp. Perhaps RNAP V-dependent regions are specifically associated with smaller repeat sequences regardless of their repeat class (). It has been demonstrated that methylation is preferentially reduced in small TEs when RdDM components AGO4 or DRM1/2 are defective, indicating that the RdDM pathway downstream of siRNA biogenesis is required to maintain the methylation state for small but not long TEs.Citation56 Therefore, it is possible that one of the key functions of RNAP V targeting in RdDM is to silence small TEs in euchromatin which are distributed too broadly and/or too small to be stably integrated into heterochromatin.Citation46,Citation52,Citation56 In the future, methylation and histone profiles of RNAP V and related mutants will allow a more detailed understanding of the function of RNAP V-dependent siRNA-directed silencing.
The current view is that DMS4, DRD1, DMS3 and RDM1 are RdDM effectors that are involved in facilitation of RNAP V transcription as RNA scaffold and recruitment of silencing complex to target genomic sites.Citation23,Citation57 Indeed, for the four RdDM effectors we examined, the degree of mutant-dependent suppression on small RNA abundance was less severe in the dms4–1, drd1–1, dms3–1 and rdm1–4 mutants compared with that of the nrpe mutant, consistent with their roles predicted to be downstream of siRNA biogenesis. Most importantly, dms3, rdm1, drd1 and dms4 were impacted predominantly at the same type of small RNA-generating regions as in nrpe, especially on small repeats of SINE and RC/Helitrons but less impacted at long repeats like LTR/Gypsy (). Our results indicated that these RdDM effectors may affect siRNA abundance mainly through their function together with RNAP V at certain RdDM target loci. However, a portion of small RNA-generating clusters in dms3, rdm1, drd1 and dms4 did not overlap with RNAP V-dependent clusters, suggesting a partially non-redundant role for these RdDM effectors on siRNA accumulation with RNAP V. One possibility is that they work together with other RNA polymerases in RdDM, for example, like DMS4 interacts with both RNAP II and RNAP V and possibly regulate their abundance and/or polymerase activity.Citation27,Citation58 The relationship between RdDM effector-dependent small RNA levels at specific genomic loci and epigenetic marks such as DNA methylation and histone modifications has yet to be fully elucidated. Future studies on these RdDM effectors will provide better understanding of their functions in small RNA-directed silencing.
Materials and Methods
Mutants and plant growth conditions
The mutant allele of nrpd1b-11 (SALK_029919), nrpd1a-4 (Salk_083051) and rdr2–1 (SAIL_1277_H08) used in this study was from Arabidopsis thaliana ecotype Columbia as described previously.Citation7,Citation16 dms4–1, dms3–1, rdm1–4 and drd1–1 were described previously.Citation22,Citation27,Citation30,Citation40 Plants were grown in a growth chamber with 16 h of light for five weeks. Immature inflorescence tissues including inflorescence meristem and early stages floral buds (up to stage 11/12) were collected. Total RNA was isolated using Tri-reagent (Molecular Research Center) according to the manufacturer’s instructions.
Small RNA data generation and analysis
Small RNA libraries were constructed as described,Citation59 and the SBS sequencing was performed on an Illumina GAIIx at the University of Delaware. The SBS data was processed and normalized as previously described.Citation60,Citation61 In brief, the raw sequencing data was converted to SCARF format, trimmed of adapters, matched to the genome (Arabidopsis TAIR version 9, “TAIR9”), and read counts normalized based on the total abundance of genome-matched small RNA reads, excluding structural sRNAs originating from annotated tRNA, rRNA, small nuclear (sn) and small nucleolar (sno) RNAs.Citation61 The “hits-normalized-abundance” (HNA) values were calculated by dividing the normalized abundance (in RP5M) for each small RNA hit, where a hit is defined as simply the number of loci at which a given sequence perfectly matches the genome. Repeat annotation was based on the repeat information in the TAIR9 genome.Citation44 These sequence data are available in GenBank’s GEO database under the accession number GSE36424.
Clustering and differential expression analysis was performed using custom Perl and database scripts.Citation61 A static clustering approach was implemented to calculate the hits-normalized abundance of all the small RNA reads in every 500 bp cluster, as described in the main text. Repeats were annotated for clusters only if more than 100 nt of the 500 nt clusters was marked as a known repeat.
To calculate the width of RNAP V dependent regions, we selected regions of total 1,500 bp; 500 bp was added both 5′ and 3′ to the selected 500 bp cluster, as described in the main text. The points in the peak represent the fold-difference between Col and nrpe (Y value) within a window of 100 bp, recalculated every 20 bp across the 1500 bp region. The peaks were smoothed to fit a Gaussian distribution to the data calculated from the moving average. To generate these graphs, a static sliding-window clustering method was implemented to re-compute these values within smaller bins of 100 bp, sliding every 20 bp across the selected 1500 bp regions. The analysis was done using custom Perl scripts and MySQL database queries, while the graphs and Gaussian distribution fits to individual peaks were generated using OriginLab software.
Additional material
Download Zip (6 MB)Acknowledgments
We gratefully acknowledge the assistance of Mayumi Nakano, Aishwarya Gurazada, and Kevin McCormick with informatics, and Bruce Kingham for sequencing. This project was supported by NSF Plant Genome Research award #0701745 to B.C.M.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Grewal SI, Jia S. Heterochromatin revisited. Nat Rev Genet 2007; 8:35 - 46; http://dx.doi.org/10.1038/nrg2008; PMID: 17173056
- Chan SW, Henderson IR, Jacobsen SE. Gardening the genome: DNA methylation in Arabidopsis thaliana. Nat Rev Genet 2005; 6:351 - 60; http://dx.doi.org/10.1038/nrg1601; PMID: 15861207
- Tamaru H. Confining euchromatin/heterochromatin territory: jumonji crosses the line. Genes Dev 2010; 24:1465 - 78; http://dx.doi.org/10.1101/gad.1941010; PMID: 20634313
- Matzke MA, Birchler JA. RNAi-mediated pathways in the nucleus. Nat Rev Genet 2005; 6:24 - 35; http://dx.doi.org/10.1038/nrg1500; PMID: 15630419
- Volpe TA, Kidner C, Hall IM, Teng G, Grewal SI, Martienssen RA. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science 2002; 297:1833 - 7; http://dx.doi.org/10.1126/science.1074973; PMID: 12193640
- Zilberman D, Cao X, Jacobsen SE. ARGONAUTE4 control of locus-specific siRNA accumulation and DNA and histone methylation. Science 2003; 299:716 - 9; http://dx.doi.org/10.1126/science.1079695; PMID: 12522258
- Xie Z, Johansen LK, Gustafson AM, Kasschau KD, Lellis AD, Zilberman D, et al. Genetic and functional diversification of small RNA pathways in plants. PLoS Biol 2004; 2:E104; http://dx.doi.org/10.1371/journal.pbio.0020104; PMID: 15024409
- Law JA, Jacobsen SE. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet 2010; 11:204 - 20; http://dx.doi.org/10.1038/nrg2719; PMID: 20142834
- Matzke M, Kanno T, Daxinger L, Huettel B, Matzke AJM. RNA-mediated chromatin-based silencing in plants. Curr Opin Cell Biol 2009; 21:367 - 76; http://dx.doi.org/10.1016/j.ceb.2009.01.025; PMID: 19243928
- Lahmy S, Bies-Etheve N, Lagrange T. Plant-specific multisubunit RNA polymerase in gene silencing. Epigenetics 2010; 5:4 - 8; http://dx.doi.org/10.4161/epi.5.1.10435; PMID: 20173420
- Pikaard CS, Haag JR, Ream T, Wierzbicki AT. Roles of RNA polymerase IV in gene silencing. Trends Plant Sci 2008; 13:390 - 7; http://dx.doi.org/10.1016/j.tplants.2008.04.008; PMID: 18514566
- Ream TS, Haag JR, Wierzbicki AT, Nicora CD, Norbeck AD, Zhu JK, et al. Subunit compositions of the RNA-silencing enzymes Pol IV and Pol V reveal their origins as specialized forms of RNA polymerase II. Mol Cell 2009; 33:192 - 203; http://dx.doi.org/10.1016/j.molcel.2008.12.015; PMID: 19110459
- Herr AJ, Jensen MB, Dalmay T, Baulcombe DC. RNA polymerase IV directs silencing of endogenous DNA. Science 2005; 308:118 - 20; http://dx.doi.org/10.1126/science.1106910; PMID: 15692015
- Onodera Y, Haag JR, Ream T, Costa Nunes P, Pontes O, Pikaard CS. Plant nuclear RNA polymerase IV mediates siRNA and DNA methylation-dependent heterochromatin formation. Cell 2005; 120:613 - 22; http://dx.doi.org/10.1016/j.cell.2005.02.007; PMID: 15766525
- Kanno T, Huettel B, Mette MF, Aufsatz W, Jaligot E, Daxinger L, et al. Atypical RNA polymerase subunits required for RNA-directed DNA methylation. Nat Genet 2005; 37:761 - 5; http://dx.doi.org/10.1038/ng1580; PMID: 15924141
- Pontier D, Yahubyan G, Vega D, Bulski A, Saez-Vasquez J, Hakimi MA, et al. Reinforcement of silencing at transposons and highly repeated sequences requires the concerted action of two distinct RNA polymerases IV in Arabidopsis. Genes Dev 2005; 19:2030 - 40; http://dx.doi.org/10.1101/gad.348405; PMID: 16140984
- Lahmy S, Pontier D, Cavel E, Vega D, El-Shami M, Kanno T, et al. PolV(PolIVb) function in RNA-directed DNA methylation requires the conserved active site and an additional plant-specific subunit. Proc Natl Acad Sci U S A 2009; 106:941 - 6; http://dx.doi.org/10.1073/pnas.0810310106; PMID: 19141635
- Li CF, Pontes O, El-Shami M, Henderson IR, Bernatavichute YV, Chan SW, et al. An ARGONAUTE4-containing nuclear processing center colocalized with Cajal bodies in Arabidopsis thaliana. Cell 2006; 126:93 - 106; http://dx.doi.org/10.1016/j.cell.2006.05.032; PMID: 16839879
- Mosher RA, Schwach F, Studholme D, Baulcombe DC. PolIVb influences RNA-directed DNA methylation independently of its role in siRNA biogenesis. Proc Natl Acad Sci U S A 2008; 105:3145 - 50; http://dx.doi.org/10.1073/pnas.0709632105; PMID: 18287047
- Wierzbicki AT, Ream TS, Haag JR, Pikaard CS. RNA polymerase V transcription guides ARGONAUTE4 to chromatin. Nat Genet 2009; 41:630 - 4; http://dx.doi.org/10.1038/ng.365; PMID: 19377477
- Zheng B, Wang Z, Li S, Yu B, Liu JY, Chen X. Intergenic transcription by RNA polymerase II coordinates Pol IV and Pol V in siRNA-directed transcriptional gene silencing in Arabidopsis. Genes Dev 2009; 23:2850 - 60; http://dx.doi.org/10.1101/gad.1868009; PMID: 19948763
- Gao Z, Liu H-L, Daxinger L, Pontes O, He X, Qian W, et al. An RNA polymerase II- and AGO4-associated protein acts in RNA-directed DNA methylation. Nature 2010; 465:106 - 9; http://dx.doi.org/10.1038/nature09025; PMID: 20410883
- Zhang H, Zhu J-K. RNA-directed DNA methylation. Curr Opin Plant Biol 2011; 14:142 - 7; http://dx.doi.org/10.1016/j.pbi.2011.02.003; PMID: 21420348
- Zheng X, Zhu J, Kapoor A, Zhu JK. Role of Arabidopsis AGO6 in siRNA accumulation, DNA methylation and transcriptional gene silencing. EMBO J 2007; 26:1691 - 701; http://dx.doi.org/10.1038/sj.emboj.7601603; PMID: 17332757
- Havecker ER, Wallbridge LM, Hardcastle TJ, Bush MS, Kelly KA, Dunn RM, et al. The Arabidopsis RNA-directed DNA methylation argonautes functionally diverge based on their expression and interaction with target loci. Plant Cell 2010; 22:321 - 34; http://dx.doi.org/10.1105/tpc.109.072199; PMID: 20173091
- Eun C, Lorkovic ZJ, Naumann U, Long Q, Havecker ER, Simon SA, et al. AGO6 functions in RNA-mediated transcriptional gene silencing in shoot and root meristems in Arabidopsis thaliana. PLoS One 2011; 6:e25730; http://dx.doi.org/10.1371/journal.pone.0025730; PMID: 21998686
- Kanno T, Bucher E, Daxinger L, Huettel B, Kreil DP, Breinig F, et al. RNA-directed DNA methylation and plant development require an IWR1-type transcription factor. EMBO Rep 2010; 11:65 - 71; http://dx.doi.org/10.1038/embor.2009.246; PMID: 20010803
- Wierzbicki AT, Haag JR, Pikaard CS. Noncoding transcription by RNA polymerase Pol IVb/Pol V mediates transcriptional silencing of overlapping and adjacent genes. Cell 2008; 135:635 - 48; http://dx.doi.org/10.1016/j.cell.2008.09.035; PMID: 19013275
- Gao Z, Liu HL, Daxinger L, Pontes O, He X, Qian W, et al. An RNA polymerase II- and AGO4-associated protein acts in RNA-directed DNA methylation. Nature 2010; 465:106 - 9; http://dx.doi.org/10.1038/nature09025; PMID: 20410883
- Kanno T, Bucher E, Daxinger L, Huettel B, Böhmdorfer G, Gregor W, et al. A structural-maintenance-of-chromosomes hinge domain-containing protein is required for RNA-directed DNA methylation. Nat Genet 2008; 40:670 - 5; http://dx.doi.org/10.1038/ng.119; PMID: 18425128
- Ausin I, Mockler TC, Chory J, Jacobsen SE. IDN1 and IDN2 are required for de novo DNA methylation in Arabidopsis thaliana. Nat Struct Mol Biol 2009; 16:1325 - 7; http://dx.doi.org/10.1038/nsmb.1690; PMID: 19915591
- He XJ, Hsu YF, Zhu S, Wierzbicki AT, Pontes O, Pikaard CS, et al. An effector of RNA-directed DNA methylation in arabidopsis is an ARGONAUTE 4- and RNA-binding protein. Cell 2009; 137:498 - 508; http://dx.doi.org/10.1016/j.cell.2009.04.028; PMID: 19410546
- Bies-Etheve N, Pontier D, Lahmy S, Picart C, Vega D, Cooke R, et al. RNA-directed DNA methylation requires an AGO4-interacting member of the SPT5 elongation factor family. EMBO Rep 2009; 10:649 - 54; http://dx.doi.org/10.1038/embor.2009.31; PMID: 19343051
- Cao X, Aufsatz W, Zilberman D, Mette MF, Huang MS, Matzke M, et al. Role of the DRM and CMT3 methyltransferases in RNA-directed DNA methylation. Curr Biol 2003; 13:2212 - 7; http://dx.doi.org/10.1016/j.cub.2003.11.052; PMID: 14680640
- Cao X, Jacobsen SE. Role of the arabidopsis DRM methyltransferases in de novo DNA methylation and gene silencing. Curr Biol 2002; 12:1138 - 44; http://dx.doi.org/10.1016/S0960-9822(02)00925-9; PMID: 12121623
- Lindroth AM, Cao X, Jackson JP, Zilberman D, McCallum CM, Henikoff S, et al. Requirement of CHROMOMETHYLASE3 for maintenance of CpXpG methylation. Science 2001; 292:2077 - 80; http://dx.doi.org/10.1126/science.1059745; PMID: 11349138
- Johnson LM, Law JA, Khattar A, Henderson IR, Jacobsen SE. SRA-domain proteins required for DRM2-mediated de novo DNA methylation. PLoS Genet 2008; 4:e1000280; http://dx.doi.org/10.1371/journal.pgen.1000280; PMID: 19043555
- Daxinger L, Kanno T, Bucher E, van der Winden J, Naumann U, Matzke AJM, et al. A stepwise pathway for biogenesis of 24-nt secondary siRNAs and spreading of DNA methylation. EMBO J 2009; 28:48 - 57; http://dx.doi.org/10.1038/emboj.2008.260; PMID: 19078964
- Zhang X, Henderson IR, Lu C, Green PJ, Jacobsen SE. Role of RNA polymerase IV in plant small RNA metabolism. Proc Natl Acad Sci U S A 2007; 104:4536 - 41; http://dx.doi.org/10.1073/pnas.0611456104; PMID: 17360559
- Kanno T, Mette MF, Kreil DP, Aufsatz W, Matzke M, Matzke AJ. Involvement of putative SNF2 chromatin remodeling protein DRD1 in RNA-directed DNA methylation. Curr Biol 2004; 14:801 - 5; http://dx.doi.org/10.1016/j.cub.2004.04.037; PMID: 15120073
- Nobuta K, Lu C, Shrivastava R, Pillay M, De Paoli E, Accerbi M, et al. Distinct size distribution of endogeneous siRNAs in maize: Evidence from deep sequencing in the mop1-1 mutant. Proc Natl Acad Sci U S A 2008; 105:14958 - 63; http://dx.doi.org/10.1073/pnas.0808066105; PMID: 18815367
- Kasschau KD, Fahlgren N, Chapman EJ, Sullivan CM, Cumbie JS, Givan SA, et al. Genome-wide profiling and analysis of Arabidopsis siRNAs. PLoS Biol 2007; 5:e57; http://dx.doi.org/10.1371/journal.pbio.0050057; PMID: 17298187
- Lu C, Kulkarni K, Souret FF, MuthuValliappan R, Tej SS, Poethig RS, et al. MicroRNAs and other small RNAs enriched in the Arabidopsis RNA-dependent RNA polymerase-2 mutant. Genome Res 2006; 16:1276 - 88; http://dx.doi.org/10.1101/gr.5530106; PMID: 16954541
- Lamesch P, Berardini TZ, Li D, Swarbreck D, Wilks C, Sasidharan R, et al. The Arabidopsis Information Resource (TAIR): improved gene annotation and new tools. Nucleic Acids Res 2012; 40:Database issue D1202 - 10; http://dx.doi.org/10.1093/nar/gkr1090; PMID: 22140109
- Smit A, Hubley R, Green P. RepeatMasker Open-3.0. <http://wwwrepeatmaskerorg> 1996-2010.
- Zilberman D, Henikoff S. Silencing of transposons in plant genomes: kick them when they’re down. Genome Biol 2004; 5:249; http://dx.doi.org/10.1186/gb-2004-5-12-249; PMID: 15575975
- Lisch D. Epigenetic regulation of transposable elements in plants. Annu Rev Plant Biol 2009; 60:43 - 66; http://dx.doi.org/10.1146/annurev.arplant.59.032607.092744; PMID: 19007329
- Goodier JL, Kazazian HH Jr.. Retrotransposons revisited: the restraint and rehabilitation of parasites. Cell 2008; 135:23 - 35; http://dx.doi.org/10.1016/j.cell.2008.09.022; PMID: 18854152
- Law JA, Ausin I, Johnson LM, Vashisht AA, Zhu JK, Wohlschlegel JA, et al. A protein complex required for polymerase V transcripts and RNA- directed DNA methylation in Arabidopsis. Curr Biol 2010; 20:951 - 6; http://dx.doi.org/10.1016/j.cub.2010.03.062; PMID: 20409711
- Copenhaver GP, Nickel K, Kuromori T, Benito MI, Kaul S, Lin X, et al. Genetic definition and sequence analysis of Arabidopsis centromeres. Science 1999; 286:2468 - 74; http://dx.doi.org/10.1126/science.286.5449.2468; PMID: 10617454
- Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 2000; 408:796 - 815; http://dx.doi.org/10.1038/35048692; PMID: 11130711
- Tran RK, Zilberman D, de Bustos C, Ditt RF, Henikoff JG, Lindroth AM, et al. Chromatin and siRNA pathways cooperate to maintain DNA methylation of small transposable elements in Arabidopsis. Genome Biol 2005; 6:R90; http://dx.doi.org/10.1186/gb-2005-6-11-r90; PMID: 16277745
- Ahmed I, Sarazin A, Bowler C, Colot V, Quesneville H. Genome-wide evidence for local DNA methylation spreading from small RNA-targeted sequences in Arabidopsis. Nucleic Acids Res 2011; 39:6919 - 31; http://dx.doi.org/10.1093/nar/gkr324; PMID: 21586580
- Huettel B, Kanno T, Daxinger L, Aufsatz W, Matzke AJ, Matzke M. Endogenous targets of RNA-directed DNA methylation and Pol IV in Arabidopsis. EMBO J 2006; 25:2828 - 36; http://dx.doi.org/10.1038/sj.emboj.7601150; PMID: 16724114
- Chodavarapu RK, Feng S, Bernatavichute YV, Chen PY, Stroud H, Yu Y, et al. Relationship between nucleosome positioning and DNA methylation. Nature 2010; 466:388 - 92; http://dx.doi.org/10.1038/nature09147; PMID: 20512117
- Tran RK, Zilberman D, de Bustos C, Ditt RF, Henikoff JG, Lindroth AM, et al. Chromatin and siRNA pathways cooperate to maintain DNA methylation of small transposable elements in Arabidopsis. Genome Biol 2005; 6:R90; http://dx.doi.org/10.1186/gb-2005-6-11-r90; PMID: 16277745
- Haag JR, Pikaard CS. Multisubunit RNA polymerases IV and V: purveyors of non-coding RNA for plant gene silencing. Nat Rev Mol Cell Biol 2011; 12:483 - 92; http://dx.doi.org/10.1038/nrm3152; PMID: 21779025
- He XJ, Hsu YF, Zhu S, Liu HL, Pontes O, Zhu J, et al. A conserved transcriptional regulator is required for RNA-directed DNA methylation and plant development. Genes Dev 2009; 23:2717 - 22; http://dx.doi.org/10.1101/gad.1851809; PMID: 19903758
- Lu C, Meyers BC, Green PJ. Construction of small RNA cDNA libraries for deep sequencing. Methods 2007; 43:110 - 7; http://dx.doi.org/10.1016/j.ymeth.2007.05.002; PMID: 17889797
- Nobuta K, McCormick K, Nakano M, Meyers BC. Bioinformatics analysis of small RNAs in plants using next generation sequencing technologies. Methods Mol Biol 2010; 592:89 - 106; http://dx.doi.org/10.1007/978-1-60327-005-2_7; PMID: 19802591
- McCormick KP, Willmann MR, Meyers BC. Experimental design, preprocessing, normalization and differential expression analysis of small RNA sequencing experiments. Silence 2011; 2:2; http://dx.doi.org/10.1186/1758-907X-2-2; PMID: 21356093
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 2003; 301:653 - 7; http://dx.doi.org/10.1126/science.1086391; PMID: 12893945