Abstract
The kinetochore is formed on centromeric DNA as a key interface with microtubules from the mitotic spindle to achieve accurate chromosome segregation during mitosis. However, in contrast to other regions of the chromosome, the position of the kinetochore is specified by sequence-independent epigenetic mechanisms. Most recent work on kinetochore specification has focused on the centromere-specific histone H3-variant CENP-A. Whereas CENP-A is an important epigenetic marker for the kinetochore specification, it is unclear how centromeric chromatin structure is organized. To understand centromeric chromatin structure, we focused on additional centromere proteins that have an intrinsic DNA binding activity and identified the DNA binding CENP-T-W-S-X complex. Tetramer formation of CENP-T-W-S-X is essential for functional kinetochore assembly in vertebrate cells. Our structural and biochemical analysis reveals that the CENP-T-W-S-X complex is composed of four histone-fold domains with structural similarity to nucleosomes and displays DNA supercoiling activity. These results suggest that the CENP-T-W-S-X complex forms a unique nucleosome-like structure at centromeric chromatin. In addition, CENP-S and CENP-X function at non-centromeric sites. The intriguing histone-like properties of these proteins suggest that they may form nucleosome-like structures at various genome loci, extending the chromatin code beyond classical histone variants.
Centromere specific histone variant CENP-A
The kinetochore is formed on centromeric DNA, but there is an ongoing debate about the contribution of DNA sequence information to kinetochore formation.Citation1 Whereas there are large arrays of repetitive sequences in human centromere regions (alphoid DNA) that contribute to the efficient construction of human artificial chromosomes,Citation2,Citation3 analysis of human neocentromeres suggests that these repetitive sequences are not absolutely essential for kinetochore formation.Citation1 Thus, the identification of neocentroneres has led to the idea that the kinetochore is specified on centromeric chromatin by sequence-independent epigenetic mechanisms. The centromere-specific histone H3-variant CENP-A is a key epigenetic marker because all active centromeres, including neocentromeres, contain CENP-A.Citation1,Citation4-Citation6 To support with this idea, several groups recently demonstrated that ectopic targeting of CENP-A to non-centromere loci induces kinetochore-like structures in vertebrate or Drosophila cells.Citation7-Citation9 Therefore, most recent studies on kinetochore specification have focused on how CENP-A is incorporated into centromeric nucleosomes.Citation4,Citation5
Constitutive Centromere Associated Network of proteins (CCAN)
Although we agree that deposition of CENP-A-containing nucleosomes is necessary for kinetochore specification, we suspected that additional components would be required to generate a centromere-specific chromatin structure and direct sequence-independent kinetochore assembly, because CENP-A is not strictly sufficient for the formation of functional kinetochores in vertebrate cells.Citation10,Citation11 To identify additional kinetochore components, we and other groups independently applied a proteomics approach and found proteins that associated with centromeric chromatin.Citation12-Citation16 Because these proteins localize to centromeres throughout the cell-cycle including during interphase, we referred them as the Constitutive Centromere Associated Network of proteins (CCAN). Currently, we have identified 16 CCAN proteins (CENP-C, -H, -I, -K, -L, -M, -N, -O, -P, -Q, -R, -S, -T, -U, -W and -X) and we have found that most CCAN proteins localize to the inner kinetochore region that corresponds to centromeric chromatin based on electron microscopy observation.Citation17
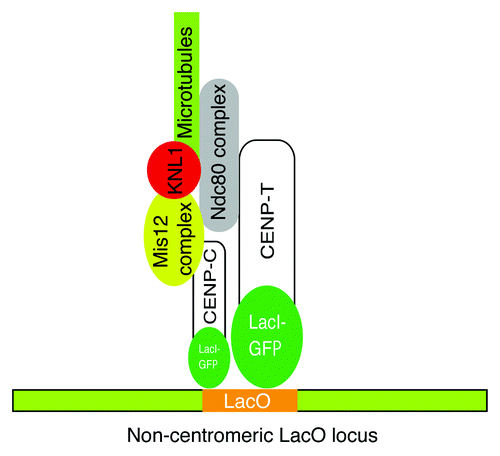
Among the CCAN proteins, we focused on CENP-T and the CENP-T associated protein CENP-W, because the CENP-T-W complex has an intrinsic DNA binding activity by which it appeared to contribute to the formation of a centromeric chromatin structure.Citation15 Interestingly, the CENP-T-W complex does not associate with CENP-A containing nucleosomes, suggesting that the complex has distinct functions from CENP-A nucleosomes to organize centromeric chromatin.Citation15,Citation18 Therefore, we hypothesized that the CENP-T-W complex (and CENP-C) serves a structural hub independently of CENP-A nucleosomes for kinetochore assembly. To test this hypothesis, we generated LacI-fusions to CENP-T or CENP-C and ectopically targeted them to a non-centromeric LacO locus in both human and chicken cells.Citation11 Consistent with our hypothesis, in each case we found that a kinetochore-like structure was formed at the LacO locus that was able to bind to spindle microtubules.Citation11 Interestingly, CENP-A was not detected at the LacO locus, indicating that ectopic localization of CENP-T (and CENP-C) induces kinetochore formation and can at least partially bypass the requirement for CENP-A nucleosomes ().Citation11
CENP-T-W-S-X Forms a Unique Nucleosome-Like Structure with a Histone Fold Domains
Using the ectopic localization experiments described above, we found that the CENP-T-W complex plays an important role for kinetochore assembly. However, little was known about how the CENP-T-W complex is targeted to centromeres and contributes to the organization centromeric-specific chromatin structure at endogenous centromeres. To address this question, we analyzed the structural, biochemical, and functional basis for association of the CENP-T-W complex with DNA.Citation19 We co-expressed the C-terminal DNA binding regions of chicken CENP-T (531–639 aa) and CENP-W. Following crystallization of the CENP-T-W complex, we determined the structure of the complex at 2.2 Å resolution. Interestingly, the structure of the CENP-T-W dimer is highly homologous to the structure of canonical histones and other histone-fold containing protein complexes, such as NC2α-β and CHRAC14–16.Citation19
In addition to the CENP-T-W complex, we have previously identified the CENP-S-X complex, which also shows sequence similarity with histones.Citation16 To test the relationship between the CENP-T-W and CENP-S-X complexes, we determined the structure of the CENP-S-X complex at 2.15 Å resolution.Citation19 Similar to the CENP-T-W complex, the structure of the CENP-S-X complex is closely related to that of canonical histones. However, the CENP-S-X complex forms a (CENP-S-X)2 tetramer, whereas the CENP-T-W complex forms a dimer.
As the overall structure of the CENP-T-W and CENP-S-X complexes is quite similar, we hypothesized that these two histone-fold containing complexes may interact. Indeed, when these complexes are combined in equimolar ratios, CENP-S-X and CENP-T-W form a single stoichiometric complex.Citation19 Based on the molecular weight of the complex, we conclude that one CENP-S-X dimer interacts with one CENP-T-W dimer to form a CENP-T-W-S-X heterotetramer. We determined the structure of the CENP-T-W-S-X complex at 2.4 Å resolution and confirmed heterotetramer formation of the CENP-T-W-S-X complex.Citation19 The structural analysis allowed us to identify amino acids responsible for the tetramer formation. To access biological significance of the tetramer formation, we generated CENP-T or CENP-S mutants that are defective for tetramer formation and introduced these mutants into CENP-T- or CENP-S-deficient cells, respectively. We found that these mutants did not localize to kinetochore properly and that a functional kinetochore was not formed in cells expressing these mutants. Thus, we conclude that tetramer formation of CENP-T-W-S-X is essential to assemble functional kinetochores.Citation19
In parallel biochemical work, we demonstrated that the CENP-T-W-S-X complex has DNA binding activity in gel shift assays. We were able to predict the DNA binding sites for the CENP-T-W-S-X heterotetramer from a structural comparison with the DNA binding surface of canonical histones. As the histone octamer wraps DNA along the surface of the nucleosome, it is possible that DNA is also bent along the surface of the CENP-T-W-S-X heterotetramer to form a nucleosome-like structure. To prove this, we demonstrated that the CENP-T-W-S-X complex has DNA supercoiling activity similar to canonical histones, which requires that DNA is wrapped along the surface.Citation19 We also demonstrated that the CENP-T-W-S-X complex formed a discrete complex with ~100 bp DNA. In addition, 100 bp DNA was protected, when the DNA-CENP-T-W-S-X complex was digested by micrococcal nuclease. Then we believe that CENP-T-W-S-X complex form a nucleosome-like structure with ~100 bp DNA. However, DNA supercoiling activity of the CENP-T-W-S-X complex is weaker than that of the canonical histone octamer, suggesting that the CENP-T-W-S-X complex wraps DNA with lower efficiency than canonical histones. Therefore, the “nucleosome-like structure” by DNA and the CENP-T-W-S-X is distinct from canonical nucleosomes.
As the CENP-T-W-S-X heterotetramer requires ~100 bp of DNA for its complete binding, it is unlikely that the CENP-T-W-S-X complex binds to chromosomal DNA in regions that are occupied by nucleosomes. Therefore, it is likely that there are nucleosome free regions at centromeres. Sun et al.Citation20 have suggested that there are nucleosome free regions of > 100 bp in eukaryotic genomes. Such ~100 bp nucleosome-free regions may be interspersed between regular nucleosomes at centromeres to provide a binding region for the CENP-T-W-S-X heterotetramer. We have previously shown that CENP-T-associated DNA isolated from cells is centromeric and the pattern of MNase digestion from CENP-T-associated chromatin is different from that of DNA isolated by CENP-A immunoprecipitations.Citation15 These data suggest the existence of a unique CENP-T-W-S-X-containing chromatin structure in vivo. We propose that there are CENP-A containing nucleosomes, canonical H3 nucleosomes, and CENP-T-W-S-X containing nucleosome-like structures interspersed in centromeric chromatin. We believe that coordination of these 3 types of structures is essential to establish a centromeric-specific chromatin structure and provide the epigenetic mark for kinetochore specification. Prendergast et al. performed the FRAP analysis on CENP-A and CENP-T and proposed that CENP-A marks the centromere, whereas CENP-T marks the kinetochores.Citation21 This proposal is consistent with our model. Obtaining definitive proof that these coordinated structures exist in vivo is a major future challenge. In addition, some modification of centromeric nucleosomes may contribute to establishment of the centromeric-specific chromatin.
A New Chromatin Code with Histone-Like Proteins
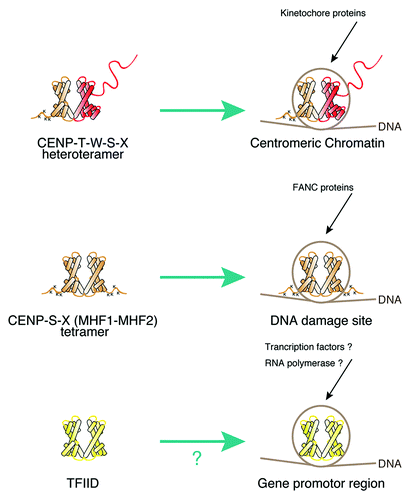
Canonical histones provide a “histone code” to specify the function of different regions of the chromosome through histone modification or multiple combinations of histone variants including the H2A variants H2AZ or macro H2A, and the H3 variants H3.3 or CENP-A.Citation22 Altering nucleosome composition using these variants allows for diverse functions. For example, nucleosomes containing H2AZ are enriched in gene promoter regionsCitation23 and CENP-A containing nucleosomes are located at centromere regions.Citation4 In addition to this canonical histone code, we propose an extended chromatin code that also involves histone-like proteins (). The CENP-T-W-S-X heterotetramer plays a key role in centromere function, but similar nucleosome-like structures may also be found at other genomic locations. Indeed, CENP-S and CENP-X are conserved kinetochore-localized proteins,Citation5,Citation16 but have also been identified as Fanconia Anemia M (FANCM) associated proteins where they were named MHF1 and MHF2. The association of the MHF1-MHF2 complex with FANCM occurs at DNA damage sites.Citation24,Citation25 At these locations, the MHF1-MHF2 (CENP-S-X) complex may act as a tetramer to associate with DNA, or may associate with additional histone fold proteins to generate distinct chromatin structures in each region (). Although there are many proteins that possess histone-fold regions including transcription factor TFIID,Citation26 the DNA binding properties of these proteins are not clear. It is possible that these proteins may also form nucleosome-like structure, which may serve a specific role in regulating transcription (). The use of nucleosome-like structures with histone-like proteins may provide a new chromatin code, which extends the canonical histone code.
Acknowledgments
The author thanks Dr. Iain Cheeseman for useful comments on the manuscript. The research in the Fukagawa Lab was supported by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan and the Cabinet Office, Government of Japan through its “Funding Program for Next Generation World-Leading Researchers.”
References
- Marshall OJ, Chueh AC, Wong LH, Choo KH. Neocentromeres: new insights into centromere structure, disease development, and karyotype evolution. Am J Hum Genet 2008; 82:261 - 82; http://dx.doi.org/10.1016/j.ajhg.2007.11.009; PMID: 18252209
- Harrington JJ, Van Bokkelen G, Mays RW, Gustashaw K, Willard HF. Formation of de novo centromeres and construction of first-generation human artificial microchromosomes. Nat Genet 1997; 15:345 - 55; http://dx.doi.org/10.1038/ng0497-345; PMID: 9090378
- Ikeno M, Grimes B, Okazaki T, Nakano M, Saitoh K, Hoshino H, et al. Construction of YAC-based mammalian artificial chromosomes. Nat Biotechnol 1998; 16:431 - 9; http://dx.doi.org/10.1038/nbt0598-431; PMID: 9592390
- Black BE, Cleveland DW. Epigenetic centromere propagation and the nature of CENP-a nucleosomes. Cell 2011; 144:471 - 9; http://dx.doi.org/10.1016/j.cell.2011.02.002; PMID: 21335232
- Perpelescu M, Fukagawa T. The ABCs of CENPs. Chromosoma 2011; 120:425 - 46; http://dx.doi.org/10.1007/s00412-011-0330-0; PMID: 21751032
- Burrack LS, Berman J. Flexibility of centromere and kinetochore structures. Trends Genet 2012; http://dx.doi.org/10.1016/j.tig.2012.02.003; PMID: 22445183
- Barnhart MC, Kuich PH, Stellfox ME, Ward JA, Bassett EA, Black BE, et al. HJURP is a CENP-A chromatin assembly factor sufficient to form a functional de novo kinetochore. J Cell Biol 2011; 194:229 - 43; http://dx.doi.org/10.1083/jcb.201012017; PMID: 21768289
- Guse A, Carroll CW, Moree B, Fuller CJ, Straight AF. In vitro centromere and kinetochore assembly on defined chromatin templates. Nature 2011; 477:354 - 8; http://dx.doi.org/10.1038/nature10379; PMID: 21874020
- Mendiburo MJ, Padeken J, Fülöp S, Schepers A, Heun P. Drosophila CENH3 is sufficient for centromere formation. Science 2011; 334:686 - 90; http://dx.doi.org/10.1126/science.1206880; PMID: 22053052
- Van Hooser AA, Ouspenski II, Gregson HC, Starr DA, Yen TJ, Goldberg ML, et al. Specification of kinetochore-forming chromatin by the histone H3 variant CENP-A. J Cell Sci 2001; 114:3529 - 42; PMID: 11682612
- Gascoigne KE, Takeuchi K, Suzuki A, Hori T, Fukagawa T, Cheeseman IM. Induced ectopic kinetochore assembly bypasses the requirement for CENP-A nucleosomes. Cell 2011; 145:410 - 22; http://dx.doi.org/10.1016/j.cell.2011.03.031; PMID: 21529714
- Obuse C, Yang H, Nozaki N, Goto S, Okazaki T, Yoda K. Proteomics analysis of the centromere complex from HeLa interphase cells: UV-damaged DNA binding protein 1 (DDB-1) is a component of the CEN-complex, while BMI-1 is transiently co-localized with the centromeric region in interphase. Genes Cells 2004; 9:105 - 20; http://dx.doi.org/10.1111/j.1365-2443.2004.00705.x; PMID: 15009096
- Okada M, Cheeseman IM, Hori T, Okawa K, McLeod IX, Yates JR 3rd, et al. The CENP-H-I complex is required for the efficient incorporation of newly synthesized CENP-A into centromeres. Nat Cell Biol 2006; 8:446 - 57; http://dx.doi.org/10.1038/ncb1396; PMID: 16622420
- Foltz DR, Jansen LE, Black BE, Bailey AO, Yates JR 3rd, Cleveland DW. The human CENP-A centromeric nucleosome-associated complex. Nat Cell Biol 2006; 8:458 - 69; http://dx.doi.org/10.1038/ncb1397; PMID: 16622419
- Hori T, Amano M, Suzuki A, Backer CB, Welburn JP, Dong Y, et al. CCAN makes multiple contacts with centromeric DNA to provide distinct pathways to the outer kinetochore. Cell 2008; 135:1039 - 52; http://dx.doi.org/10.1016/j.cell.2008.10.019; PMID: 19070575
- Amano M, Suzuki A, Hori T, Backer C, Okawa K, Cheeseman IM, et al. The CENP-S complex is essential for the stable assembly of outer kinetochore structure. J Cell Biol 2009; 186:173 - 82; http://dx.doi.org/10.1083/jcb.200903100; PMID: 19620631
- Suzuki A, Hori T, Nishino T, Usukura J, Miyagi A, Morikawa K, et al. Spindle microtubules generate tension-dependent changes in the distribution of inner kinetochore proteins. J Cell Biol 2011; 193:125 - 40; http://dx.doi.org/10.1083/jcb.201012050; PMID: 21464230
- Ribeiro SA, Vagnarelli P, Dong Y, Hori T, McEwen BF, Fukagawa T, et al. A super-resolution map of the vertebrate kinetochore. Proc Natl Acad Sci U S A 2010; 107:10484 - 9; http://dx.doi.org/10.1073/pnas.1002325107; PMID: 20483991
- Nishino T, Takeuchi K, Gascoigne KE, Suzuki A, Hori T, Oyama T, et al. CENP-T-W-S-X forms a unique centromeric chromatin structure with a histone-like fold. Cell 2012; 148:487 - 501; http://dx.doi.org/10.1016/j.cell.2011.11.061; PMID: 22304917
- Sun W, Xie W, Xu F, Grunstein M, Li KC. Dissecting nucleosome free regions by a segmental semi-Markov model. PLoS One 2009; 4:e4721; http://dx.doi.org/10.1371/journal.pone.0004721; PMID: 19266098
- Prendergast L, van Vuuren C, Kaczmarczyk A, Doering V, Hellwig D, Quinn N, et al. Premitotic assembly of human CENPs -T and -W switches centromeric chromatin to a mitotic state. PLoS Biol 2011; 9:e1001082; http://dx.doi.org/10.1371/journal.pbio.1001082; PMID: 21695110
- Jenuwein T, Allis CD. Translating the histone code. Science 2001; 293:1074 - 80; http://dx.doi.org/10.1126/science.1063127; PMID: 11498575
- Albert I, Mavrich TN, Tomsho LP, Qi J, Zanton SJ, Schuster SC, et al. Translational and rotational settings of H2A.Z nucleosomes across the Saccharomyces cerevisiae genome. Nature 2007; 446:572 - 6; http://dx.doi.org/10.1038/nature05632; PMID: 17392789
- Yan Z, Delannoy M, Ling C, Daee D, Osman F, Muniandy PA, et al. A histone-fold complex and FANCM form a conserved DNA-remodeling complex to maintain genome stability. Mol Cell 2010; 37:865 - 78; http://dx.doi.org/10.1016/j.molcel.2010.01.039; PMID: 20347428
- Singh TR, Saro D, Ali AM, Zheng XF, Du CH, Killen MW, et al. MHF1-MHF2, a histone-fold-containing protein complex, participates in the Fanconi anemia pathway via FANCM. Mol Cell 2010; 37:879 - 86; http://dx.doi.org/10.1016/j.molcel.2010.01.036; PMID: 20347429
- Gangloff YG, Romier C, Thuault S, Werten S, Davidson I. The histone fold is a key structural motif of transcription factor TFIID. Trends Biochem Sci 2001; 26:250 - 7; http://dx.doi.org/10.1016/S0968-0004(00)01741-2; PMID: 11295558