Abstract
The micronutrients folate and selenium may modulate DNA methylation patterns by affecting intracellular levels of the methyl donor S-adenosylmethionine (SAM) and/or the product of methylation reactions S-adenosylhomocysteine (SAH). WI-38 fibroblasts and FHC colon epithelial cells were cultured in the presence of two forms of folate or four forms of selenium at physiologically-relevant doses, and their effects on LINE-1 methylation, gene-specific CpG island (CGI) methylation and intracellular SAM:SAH were determined. At physiologically-relevant doses the forms of folate or selenium had no effect on LINE-1 or CGI methylation, nor on intracellular SAM:SAH. However the commercial cell culture media used for the selenium studies, containing supra-physiological concentrations of folic acid, induced LINE-1 hypomethylation, CGI hypermethylation and decreased intracellular SAM:SAH in both cell lines. We conclude that the exposure of normal human cells to supra-physiological folic acid concentrations present in commercial cell culture media perturbs the intracellular SAM:SAH ratio and induces aberrant DNA methylation.
Introduction
Altered patterns of DNA methylation are frequently observed in many human diseases, including most cancers and cardiovascular disease.Citation1-Citation3 These changes include both a general loss of methylation from the genome (global hypomethylation), which has been suggested to induce genomic instability, and gene-specific CpG island hypermethylation that often induces transcriptional silencing of associated genes.Citation1 Aberrant DNA methylation changes also occur progressively with age in many apparently healthy tissues in the human body, where it has been proposed to contribute to disease vulnerability.Citation4,Citation5
Epidemiologic studies suggest that the micronutrients folate and selenium may modulate the risk of several human diseases.Citation6-Citation10 The main biochemical role for folate is in the supply of one-carbon moieties for DNA synthesis and the synthesis of S-adenosylmethionine (SAM), the universal methyl donor in biological methylation reactions including the methylation of DNA.Citation11,Citation12 Selenium also plays a role in the metabolism of SAM.Citation13 Many studies have, therefore, investigated the impact of modulating folate and/or selenium levels on DNA methylation in vitro and in vivo with equivocal results that may reflect differences in cellular or animal models and the use of different forms of the micronutrients.Citation14,Citation15
In this study we investigated the impact on DNA methylation of two nutritionally-relevant forms of folate; folic acid used in supplements and fortified foods, and 5-methyltetrahydrofolate the naturally occurring form, and four forms of selenium; sodium selenite, selenomethionine, selenium methylselenocysteine or methylselininic acid, all at physiologically-relevant doses in two non-transformed cell lines. At these doses neither form of folate nor any form of selenium had a significant effect on either global DNA or gene-specific, CpG island methylation. However the selenium investigations were conducted in recommended cell culture media containing supra-physiological (≥ 1µM) concentrations of folic acid, which induced global DNA hypomethylation and CpG island hypermethylation in both cell lines and decreased the intracellular ratio of SAM:SAH.
Results
The effects of physiologically-relevant doses of two forms of folate; folic acid and 5-methyltetrahydrofolate, and four forms of selenium; selenite, selenomethionine, selenium methylselenocysteine and methylseleninic acid, on global and gene-specific DNA methylation were investigated in the non-transformed human cell lines WI-38 lung fibroblasts and FHC fetal colonic epithelial cells, cultured over an extended period of three passages (calculated as approximately 17 and 20 population doublings for WI-38 and FHC cells, respectively). There were no statistically significant effects of form or dose for either of the micronutrients (data not shown) suggesting that at physiologically-relevant doses neither form of folate, nor any form of selenium impacted upon DNA methylation patterns in the two normal cell lines. However, significantly different results were observed between the two sets of experiments including a time-dependent decrease in LINE-1 methylation and a time-dependent increase in gene-specific, CpG island methylation that were consistent in both cell lines across all forms and doses of selenium,but was not observed in either cell line in the experiments with physiologically-relevant levels of folic acid/metafolin. Since the major difference between the experiments was the media composition (see Table S1), in particular the level of folate, which was in the nanomolar range for the investigation of folate (using supplemented folate-free RPMI 1640 medium) but in the micromolar range for the investigation of selenium (using EMEM and DMEM media), the data from each of the experiments were pooled into physiologically “normal” (4.5 – 68 nM), i.e., representative of the normal range of human plasma folate levels,Citation16 and “supra-physiological” (2.3 – 9.1 μM) folate groups for further analysis.
These untransformed cell lines have a limited proliferative capacity in vitro and eventually become senescent after prolonged time in culture. We investigated the possibility that senescence may have been induced by quantifying senescence-associated β-Galactosidase activityCitation17 in cells at each passage but this remained undetectable throughout the experiments (data not shown) indicating that senescence was not induced in either cell line even after 3 passages.
Genomic DNA methylation
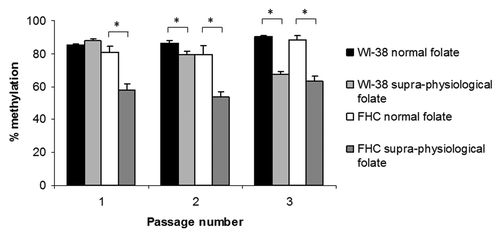
Normal folate levels had no significant effect on LINE-1 methylation, used here as a surrogate for genomic DNA methylation,Citation18 in either WI-38 or FHC cells (). However, supra-physiological folate concentrations significantly (p < 0.05) decreased LINE-1 methylation in both cell lines. LINE-1 methylation in WI-38 cells exposed to supra-physiological folate concentrations continued to decrease during time in culture (p < 0.005) (). In FHC cells supra-physiological folate concentrations induced a significant loss of LINE-1 methylation at passage 1 (p < 0.001) but no further decrease in genomic DNA methylation was observed with continued time in culture ().
Figure 1. LINE-1 methylation in normal human fibroblasts (WI-38) and colon epithelial cells (FHC) exposed to normal (n = 12) or supra-physiological (n = 18) folate concentrations over 3 passages. Columns and bars show the mean ± standard error. *p < 0.05.
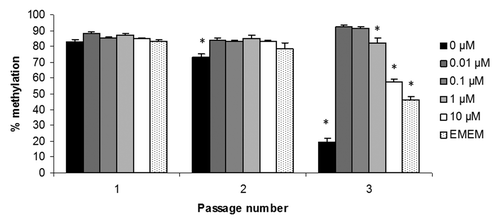
To confirm that the effects on LINE-1 methylation were induced by supra-physiological concentrations of folate and not by other minor differences in media composition between RPMI 1640 and EMEM/DMEM, WI-38 fibroblasts were cultured in RPMI 1640 supplemented with 0 to 10 µM folic acid or EMEM as before. LINE-1 methylation was quantified in genomic DNA extracted from cells obtained at each passage. shows that at passage 2, folate deficiency (0 µM) decreased LINE-1 methylation significantly (p < 0.05), which decreased further at passage 3 (p < 0.001). Folic acid concentrations of 1 µM and above also induced significant (p < 0.05) LINE-1 hypomethylation confirming that the previous observations on DNA methylation were due to the supra-physiological folic acid concentrations present in the EMEM (2.3 µM) and DMEM (9.1 µM).
Gene-specific CpG island methylation
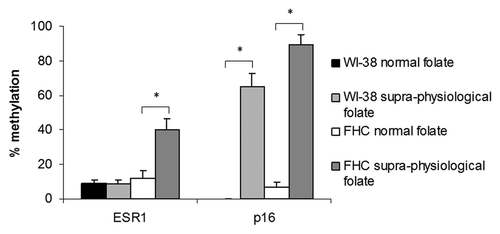
CpG island methylation was measured for the genes ESR1 and p16 in WI-38 and FHC cells after 3 passages in media containing normal (RPMI) or supra-physiological (EMEM/DMEM) folate concentrations. Folate concentration had no significant effect on ESR1 methylation in WI-38 cells; however supra-physiological folate concentrations induced significant ESR1 hypermethylation in FHC cells (p < 0.005) (). Significant hypermethylation of the p16 CpG island was also induced by supra-physiological folate concentrations in both normal cell lines (p < 0.001) ().
SAM:SAH ratio
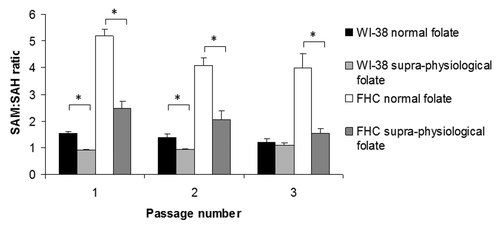
The intracellular concentrations of SAM and SAH were determined in both cell lines after each passage in media containing normal (RPMI) or supra-physiological (EMEM/DMEM) levels of folate and used to calculate the ratio of SAM to SAH, which is considered an indicator of cellular methylation potential.Citation19 shows that treatment with supra-physiological folate concentrations compared with normal folate concentrations significantly (p < 0.001) decreased the SAM:SAH ratio in both cell lines at each passage except in WI-38 cells at passage 3 where the decrease was not statistically significant.
Discussion
In the present investigation we have shown that physiologically-relevant concentrations of two forms of folate and four forms of selenium did not modulate DNA methylation patterns in normal human cell lines. However, significant differences in DNA methylation patterns were observed between the experiments with folate and those with selenium. Further analysis showed that the time-dependent induction of aberrant DNA methylation, including decreased LINE-1 methylation and increased gene-specific CpG island methylation, was due to the presence of supra-physiological folic acid concentrations in the culture media used in the experiments with the selenium compounds. The induction of aberrant DNA methylation by supra-physiological folic acid concentrations was associated with a decreased SAM:SAH ratio in both cell lines, suggesting that this perturbed cellular ‘methylation potential’ may, at least in part, be responsible.
Mathematical modeling of one-carbon metabolism supports the possibility that increased availability of folate might ‘drive’ DNA methylation through an increased intracellular supply of SAM.Citation20 Our observations that exposure to high levels of folic acid led to a decreased SAM:SAH ratio and global DNA hypomethylation suggests that excess folic acid may actually decrease the folate pool (defined as intracellular folate-related substrates e.g., tetrahydrofolate, 5-methyltetrahydrofolate, etc.), perhaps via an inhibition of many of the folate-requiring enzymesCitation21 or via as yet unknown effects of intracellular, unmetabolised folic acid. The model of Nijhout et al.Citation20 also predicted the DNA methylation reaction rate based on the kinetic characteristics of the maintenance DNA methyltransferase (DNMT1) and assumed that the other methyltransferases behaved similarly and that the DNMT pool remained constant. However, recent data demonstrated that the expression of the de novo methyltransferases DNMT3A and 3B were significantly increased in glioma cells in response to folic acid supplementation.Citation22 Together with the observation that overexpression of Dnmt3b in the murine colon led to the induction of aberrant CpG island methylation,Citation23 this suggests that cellular exposure to supra-physiological folic acid concentrations induced aberrant CpG island methylation by increasing the expression of the de novo methyltransferases DNMT3A and 3B. However this conjecture remains to be established.
While the doses of folic acid required to induce aberrant DNA methylation over the timeframe of these experiments are not nutritionally relevant to humans, these observations raise the possibility that prolonged exposure to increased, supra-nutritional doses of folic acid through the consumption of supplements or fortified foods over a life time may also induce aberrant DNA methylation. This is consistent with observations from recent studies showing a positive association between increased folate status and increased gene-specific, CpG island methylation in the normal colonic mucosa of human subjects,Citation24 (and Belshaw in preparation). These observations provide support for concerns raised recently that the provision of excessive levels of folate through mandatory food fortification with folic acid may promote the development of diseases such as cancerCitation25 via mechanisms that may include induced aberrant DNA methylation, and indicate that further studies are urgently required to understand this relationship.
There are numerous reports in the literature describing the effects of various factors on DNA methylation in cells cultured in standard commercial media, which in the vast majority of cases contain folic acid concentrations in excess of 1 µM.Citation26 Our observations, therefore, raise concerns that the results of such studies must be considered in the context that the cells are under non-physiological ‘stress’ induced by high levels of folate, and caution should be exercised in their interpretation.
Previous in vitro studies on the effects of folate on DNA methylation have utilized transformed or immortalized cell lines, which already exhibit the aberrant DNA methylation patterns we show to be induced by folic acid in the normal, non-transformed cell lines. Also, since the effects of folic acid were very similar in both cell lines, each representing very different cell types, fibroblasts and epithelial cells, our results are consistent with the possibility that the effects of excess folic acid on DNA methylation are generic across a range of tissues. Indeed, similar effects of prolonged culture on DNA methylation have been reported previously for embryonic stem cells, where the induced changes indicated they may no longer be of use in in vivo therapeutic applications.Citation27 Our observations suggest that decreasing the folic acid content of cell culture media to more physiologically-relevant levels warrants further investigation for maintaining DNA methylation patterns in these cells.
Global DNA hypomethylation, concomitant with hypermethylation of CpG islands associated with a number of genes including ESR1 and p16, has been reported to be a feature of aging in a number of human tissues.Citation5,Citation28-Citation30 Indeed, cultured fibroblast cell lines such as WI-38 have been used as models for aging in a number of studies.Citation31 However, all these studies used standard culture media, such as EMEM and DMEM. Our observations suggest that the aberrant methylation patterns observed in such studies were induced by high levels of folic acid in the culture media. It remains to be established whether the accelerated aging-related changes in DNA methylation patterns induced in cells in culture by supra-physiological folic acid concentrations completely parallel the molecular changes induced by normal aging in human tissues.
In addition to the global hypomethylating effect of supra-physiological concentrations of folic acid, we also observed significantly decreased LINE-1 methylation in WI-38 cells exposed to folate-free media. This is consistent with previous studies both in vitro and in vivo showing the induction of global hypomethylation upon folate depletion.Citation14 Together with our observed effects of supra-physiological concentrations of folic acid, this suggests the existence of an optimal range of folate concentrations for preventing effects on DNA methylation, and that both deficiency and excess may induce these adverse effects.
In conclusion, extended culture of normal human cells in physiologically-relevant levels of folate maintained normal patterns of DNA methylation. However, exposure of cells to the high levels of folic acid present in most commercial cell culture media induced patterns of both global hypomethylation and CpG island hypermethylation, similar to those observed in aged or diseased human tissues. Further studies investigating the mechanisms of folate-induced aberrant DNA methylation and the consequences for disease risk are warranted.
Materials and Methods
Cell lines and cell culture
The human lung fibroblast cell line WI-38 (ATCC CCL-75) and the colon epithelial cell line FHC (ATCC CRL-1831) were cultured in monolayers in media described below, supplemented with antibiotics (100 U/ml penicillin and 100 μg/ml streptomycin; Sigma-Aldrich) at 37°C in humidified air with 5% CO2. For the investigation of dose and chemical form of folate, both WI-38 and FHC cell lines were cultured in folate-free RPMI 1640 medium (Invitrogen) containing 10% defined fetal bovine serum (dFBS) (containing 8.8 ng/ml folate, 29 ng/ml selenium; HyClone, Thermo Scientific, Utah) supplemented with either folic acid (Sigma-Aldrich) or 5-methyltetrahydrofolate (Metafolin®; Merck) at 4.5, 18 and 68 nM, equivalent to human plasma folate status representative of deficiency, sufficiency and toward the upper limit.Citation16,Citation32
For the investigation into the effects of dose and chemical form of selenium, WI-38 and FHC cells were cultured in EMEM (2.3 µM folic acid) and DMEM (9.1 µM folic acid) media (Sigma-Aldrich), respectively, with the addition of 2.5% dFBS and supplemented with sodium selenite, selenomethionine, selenium methylselenocysteine or methylselininic acid (Sigma-Aldrich) at 0, 0.5 or 2 μM, representing deficiency through to the upper normal range.Citation33 Culture media were changed twice per week and cells were grown until they reached approximately 70% confluence to avoid potential confluence-induced effects on DNA methylation as reported previously.Citation34 Cells were harvested and passaged following treatment with 0.5% trypsin, 0.5 mM EDTA solution (Sigma-Aldrich).
DNA methylation analyses
Genomic DNA was extracted from the cells by incubation in 0.2 M TRIS-HCl pH 8.0, 0.25 M NaCl, 25 mM EDTA, 0.5% SDS containing 1 mg/ml proteinase K at 60°C for 16 h, ethanol precipitated, and resuspended in 10mM TRIS-HCl pH 8.5. The genomic DNA was converted with bisulfite as described previously.Citation35 LINE-1 methylation, a surrogate for genomic DNA methylation statusCitation18 was measured using a quantitative-PCR approach with primers described previously.Citation36 Gene-specific CpG island methylation was measured for the genes for estrogen receptor α (ESR1) and cyclin-dependent kinase inhibitor 2A (p16) using a quantitative methylation-specific PCR assay with primers described previously.Citation5 These genes were selected as they have been shown previously to be subject to aberrant methylation during both aging and disease in a number of different tissues.Citation5,Citation28-Citation30
Determination of intracellular SAM and SAH concentrations
Intracellular SAM and SAH concentrations were determined by HPLC using the method of Wang et al.Citation37
Statistical analysis
Data were analyzed using ANOVA. If a significant result was obtained (p < 0.05), further analysis was undertaken using post-hoc pairwise comparisons (Tukey HSD test). Data was subjected to diagnostic checks (e.g., outlier removal, normality of residuals) as part of the analysis process. For comparisons between the effects of normal vs. supra-physiological concentrations of folic acid, data from cells grown in RPMI supplemented with folic acid or metafolin at physiologically-relevant doses were pooled as ‘normal folate’ and data from cells grown in EMEM (WI-38) or DMEM (FHC) supplemented with selenium at physiologically-relevant doses were pooled as ‘supra-physiological folate.’ All comparisons were corrected for multiple testing using the Dunnett’s post-hoc test.
| Abbreviations: | ||
| SAM | = | S-adenosylmethionine |
| SAH | = | S-adenosylhomocysteine |
| dFBS | = | defined fetal bovine serum |
Additional material
Download Zip (40.9 KB)Acknowledgments
This work was supported by a BBSRC-funded PhD studentship to M.A.C. and by the BBSRC’s core strategic grant to the Institute of Food Research (I.T.J. and N.J.B.). We also thank Jack Dainty for his support with statistical analysis.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Author contribution
N.J.B. and I.T.J. conceived the project, developed the research plan and oversaw the research; M.A.C. conducted the research and collected the data. M.A.C. and N.J.B. analyzed the data and performed statistical analysis. M.A.C., I.T.J. and N.J.B. wrote the paper. N.J.B. had primary responsibility for final content. All authors read and approved the final manuscript.
References
- Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet 2002; 3:415 - 28; PMID: 12042769
- Lorenzen JM, Martino F, Thum T. Epigenetic modifications in cardiovascular disease. Basic Res Cardiol 2012; 107:245; http://dx.doi.org/10.1007/s00395-012-0245-9; PMID: 22234702
- Robertson KD. DNA methylation and human disease. Nat Rev Genet 2005; 6:597 - 610; http://dx.doi.org/10.1038/nrg1655; PMID: 16136652
- Christensen BC, Houseman EA, Marsit CJ, Zheng S, Wrensch MR, Wiemels JL, et al. Aging and environmental exposures alter tissue-specific DNA methylation dependent upon CpG island context. PLoS Genet 2009; 5:e1000602; http://dx.doi.org/10.1371/journal.pgen.1000602; PMID: 19680444
- Belshaw NJ, Elliott GO, Foxall RJ, Dainty JR, Pal N, Coupe A, et al. Profiling CpG island field methylation in both morphologically normal and neoplastic human colonic mucosa. Br J Cancer 2008; 99:136 - 42; http://dx.doi.org/10.1038/sj.bjc.6604432; PMID: 18542073
- Kune G, Watson L. Colorectal cancer protective effects and the dietary micronutrients folate, methionine, vitamins B6, B12, C, E, selenium, and lycopene. Nutr Cancer 2006; 56:11 - 21; http://dx.doi.org/10.1207/s15327914nc5601_3; PMID: 17176213
- Reid ME, Duffield-Lillico AJ, Slate E, Natarajan N, Turnbull B, Jacobs E, et al. The nutritional prevention of cancer: 400 mcg per day selenium treatment. Nutr Cancer 2008; 60:155 - 63; http://dx.doi.org/10.1080/01635580701684856; PMID: 18444146
- Wang X, Qin X, Demirtas H, Li J, Mao G, Huo Y, et al. Efficacy of folic acid supplementation in stroke prevention: a meta-analysis. Lancet 2007; 369:1876 - 82; http://dx.doi.org/10.1016/S0140-6736(07)60854-X; PMID: 17544768
- Rayman MP. Selenium and human health. Lancet 2012; 379:1256 - 68
- McNulty H, Pentieva K, Hoey L, Ward M. Homocysteine, B-vitamins and CVD. Proc Nutr Soc 2008; 67:232 - 7; http://dx.doi.org/10.1017/S0029665108007076; PMID: 18412997
- Stempak JM, Sohn KJ, Chiang EP, Shane B, Kim YI. Cell and stage of transformation-specific effects of folate deficiency on methionine cycle intermediates and DNA methylation in an in vitro model. Carcinogenesis 2005; 26:981 - 90; http://dx.doi.org/10.1093/carcin/bgi037; PMID: 15695236
- Sibani S, Melnyk S, Pogribny IP, Wang W, Hiou-Tim F, Deng L, et al. Studies of methionine cycle intermediates (SAM, SAH), DNA methylation and the impact of folate deficiency on tumor numbers in Min mice. Carcinogenesis 2002; 23:61 - 5; http://dx.doi.org/10.1093/carcin/23.1.61; PMID: 11756224
- Uthus EO, Ross SA, Davis CD. Differential effects of dietary selenium (se) and folate on methyl metabolism in liver and colon of rats. Biol Trace Elem Res 2006; 109:201 - 14; http://dx.doi.org/10.1385/BTER:109:3:201; PMID: 16632891
- Crider KS, Yang TP, Berry RJ, Bailey LB. Folate and DNA methylation: a review of molecular mechanisms and the evidence for folate’s role. Adv Nutr 2012; 3:21 - 38; PMID: 22332098
- Park LK, Friso S, Choi SW. Nutritional influences on epigenetics and age-related disease. Proc Nutr Soc 2012; 71:75 - 83; http://dx.doi.org/10.1017/S0029665111003302; PMID: 22051144
- de Bree A, van Dusseldorp M, Brouwer IA, van het Hof KH, Steegers-Theunissen RP. Folate intake in Europe: recommended, actual and desired intake. Eur J Clin Nutr 1997; 51:643 - 60; http://dx.doi.org/10.1038/sj.ejcn.1600467; PMID: 9347284
- Gary RK, Kindell SM. Quantitative assay of senescence-associated beta-galactosidase activity in mammalian cell extracts. Anal Biochem 2005; 343:329 - 34; http://dx.doi.org/10.1016/j.ab.2005.06.003; PMID: 16004951
- Weisenberger DJ, Campan M, Long TI, Kim M, Woods C, Fiala E, et al. Analysis of repetitive element DNA methylation by MethyLight. Nucleic Acids Res 2005; 33:6823 - 36; http://dx.doi.org/10.1093/nar/gki987; PMID: 16326863
- Caudill MA, Wang JC, Melnyk S, Pogribny IP, Jernigan S, Collins MD, et al. Intracellular S-adenosylhomocysteine concentrations predict global DNA hypomethylation in tissues of methyl-deficient cystathionine beta-synthase heterozygous mice. J Nutr 2001; 131:2811 - 8; PMID: 11694601
- Nijhout HF, Reed MC, Anderson DF, Mattingly JC, James SJ, Ulrich CM. Long-range allosteric interactions between the folate and methionine cycles stabilize DNA methylation reaction rate. Epigenetics 2006; 1:81 - 7; http://dx.doi.org/10.4161/epi.1.2.2677; PMID: 17998813
- Nijhout HF, Reed MC, Budu P, Ulrich CM. A mathematical model of the folate cycle: new insights into folate homeostasis. J Biol Chem 2004; 279:55008 - 16; http://dx.doi.org/10.1074/jbc.M410818200; PMID: 15496403
- Hervouet E, Debien E, Campion L, Charbord J, Menanteau J, Vallette FM, et al. Folate supplementation limits the aggressiveness of glioma via the remethylation of DNA repeats element and genes governing apoptosis and proliferation. Clin Cancer Res 2009; 15:3519 - 29; http://dx.doi.org/10.1158/1078-0432.CCR-08-2062; PMID: 19451595
- Steine EJ, Ehrich M, Bell GW, Raj A, Reddy S, van Oudenaarden A, et al. Genes methylated by DNA methyltransferase 3b are similar in mouse intestine and human colon cancer. J Clin Invest 2011; 121:1748 - 52; http://dx.doi.org/10.1172/JCI43169; PMID: 21490393
- Wallace K, Grau MV, Levine AJ, Shen L, Hamdan R, Chen X, et al. Association between folate levels and CpG Island hypermethylation in normal colorectal mucosa. Cancer Prev Res (Phila) 2010; 3:1552 - 64; http://dx.doi.org/10.1158/1940-6207.CAPR-10-0047; PMID: 21149331
- Smith AD, Kim YI, Refsum H. Is folic acid good for everyone?. Am J Clin Nutr 2008; 87:517 - 33; PMID: 18326588
- Johnson IT, Belshaw NJ. Environment, diet and CpG island methylation: epigenetic signals in gastrointestinal neoplasia. Food Chem Toxicol 2008; 46:1346 - 59; http://dx.doi.org/10.1016/j.fct.2007.09.101; PMID: 17976883
- Maitra A, Arking DE, Shivapurkar N, Ikeda M, Stastny V, Kassauei K, et al. Genomic alterations in cultured human embryonic stem cells. Nat Genet 2005; 37:1099 - 103; http://dx.doi.org/10.1038/ng1631; PMID: 16142235
- Issa JP. Age-related epigenetic changes and the immune system. Clin Immunol 2003; 109:103 - 8; http://dx.doi.org/10.1016/S1521-6616(03)00203-1; PMID: 14585281
- Issa JP, Ottaviano YL, Celano P, Hamilton SR, Davidson NE, Baylin SB. Methylation of the oestrogen receptor CpG island links ageing and neoplasia in human colon. Nat Genet 1994; 7:536 - 40; http://dx.doi.org/10.1038/ng0894-536; PMID: 7951326
- Post WS, Goldschmidt-Clermont PJ, Wilhide CC, Heldman AW, Sussman MS, Ouyang P, et al. Methylation of the estrogen receptor gene is associated with aging and atherosclerosis in the cardiovascular system. Cardiovasc Res 1999; 43:985 - 91; http://dx.doi.org/10.1016/S0008-6363(99)00153-4; PMID: 10615426
- Phipps SM, Berletch JB, Andrews LG, Tollefsbol TO. Aging cell culture: methods and observations. Methods Mol Biol 2007; 371:9 - 19; http://dx.doi.org/10.1007/978-1-59745-361-5_2; PMID: 17634570
- Pfeiffer CM, Caudill SP, Gunter EW, Osterloh J, Sampson EJ. Biochemical indicators of B vitamin status in the US population after folic acid fortification: results from the National Health and Nutrition Examination Survey 1999-2000. Am J Clin Nutr 2005; 82:442 - 50; PMID: 16087991
- Van Cauwenbergh R, Robberecht H, Van Vlaslaer V, Deelstra H. Comparison of the serum selenium content of healthy adults living in the Antwerp region (Belgium) with recent literature data. J Trace Elem Med Biol 2004; 18:99 - 112; http://dx.doi.org/10.1016/j.jtemb.2004.04.004; PMID: 15487770
- Pieper RO, Lester KA, Fanton CP. Confluence-induced alterations in CpG island methylation in cultured normal human fibroblasts. Nucleic Acids Res 1999; 27:3229 - 35; http://dx.doi.org/10.1093/nar/27.15.3229; PMID: 10454622
- Belshaw NJ, Elliott GO, Williams EA, Bradburn DM, Mills SJ, Mathers JC, et al. Use of DNA from human stools to detect aberrant CpG island methylation of genes implicated in colorectal cancer. Cancer Epidemiol Biomarkers Prev 2004; 13:1495 - 501; PMID: 15342451
- Iacopetta B, Grieu F, Phillips M, Ruszkiewicz A, Moore J, Minamoto T, et al. Methylation levels of LINE-1 repeats and CpG island loci are inversely related in normal colonic mucosa. Cancer Sci 2007; 98:1454 - 60; http://dx.doi.org/10.1111/j.1349-7006.2007.00548.x; PMID: 17640302
- Wang W, Kramer PM, Yang S, Pereira MA, Tao L. Reversed-phase high-performance liquid chromatography procedure for the simultaneous determination of S-adenosyl-L-methionine and S-adenosyl-L-homocysteine in mouse liver and the effect of methionine on their concentrations. J Chromatogr B Biomed Sci Appl 2001; 762:59 - 65; http://dx.doi.org/10.1016/S0378-4347(01)00341-3; PMID: 11589459