Abstract
In the developing forebrain, neural stem and progenitor cells generate a large variety of neurons with specific functions in the mature cortex. A central issue is to understand the roles of transcriptional networks and regulatory pathways that control these complex developmental processes. The proto-oncogene Ski is a transcriptional regulator linked to the human 1p36 deletion syndrome, which involves a set of phenotypes including nervous system defects. Ski shows a dynamic expression pattern during cortical development and, accordingly, the phenotype of Ski-deficient cortices is complex, involving altered cell cycle characteristics of neural progenitors, disturbed timing of neurogenesis and misspecification of projection neurons. Ski is likely to play a role in various pathways by virtue of its ability to interact with a range of signaling molecules, thereby modulating transcriptional activity of corresponding target genes. Ski regulates proliferation and differentiation of various cell types, and more recent data from my laboratory demonstrates that Ski is also involved in the specification of cortical projection neurons. This Point-of-View elucidates the role of Ski as an essential linker between sequence-specific transcription factors and non-DNA binding cofactors with chromatin modifying activities. In particular, it puts forward the hypothesis that the diverse functions of Ski as a co-repressor might be related to its association with distinct HDAC-complexes.
The transcriptional regulator Ski binds DNA only in association with other proteins.Citation1 It contains several structural domains, such as the Dachshund homology domain (DHD)Citation2 and the SAND domain (named after Sp100, AIRE-1, NucP41/75 and DEAF-1), which mediate protein-protein interactions. The SAND domain is a sequence motif found in a number of nuclear proteins that are involved in chromatin-dependent transcriptional regulation.Citation3 First evidence that Ski might be involved in epigenetic processes came from studies in Xenopus and zebrafish, where Ski transcripts and protein were found to be maternally regulated.Citation4-Citation6 Indeed, subsequent experiments revealed that Ski forms complexes with class I histone deacetylases (HDAC1–3),Citation7,Citation8 which contribute to epigenetic gene silencing by removing acetyl modifications from histones.
Ski and the Sin3/HDAC Repressor Complex
Class I HDACs are part of two major multisubunit protein assemblies, the NuRD (nucleosome remodeling and deacetylase repressor) complex and the Sin3 complex [here including the closely related N-CoR (nuclear receptor co-repressor) and SMRT (silencing mediator for retinoid and thyroid hormone receptors) co-repressors].Citation9 Initial yeast two-hybrid screens identified mSin3A, N-CoR and SMRT as direct interacting partners of Ski, defining Ski as a component of the Sin3/HDAC complex.Citation7 This was further confirmed by the finding that Ski interacts with the methyl-CpG-binding protein MeCP2, which associates with mSin3 and N-CoR.Citation10 In addition to binding to Sin3/HDAC co-repressors, Ski physically interacts with sequence-specific transcription factors that associate with the Sin3/HDAC complex. These include members of the Smad family, tumor suppressors and hormone receptors.Citation11 Notably, the described interactions of Ski with components of the Sin3/HDAC and their associated transcription factors were detected in cell lines overexpressing Ski and in cancer cells where Ski protein levels are highly elevated.Citation11 In these paradigms, loss of function studies repeatedly demonstrated that in the absence of Ski, HDAC is no longer recruited to the Sin3 repressor complex leading to abrogation of the transcriptional repression of target genes. Thus, Ski appears to serve as a scaffold protein that bridges connections between HDAC, Sin3 and cell-type specific transcription factors. There is increasing evidence that Sin3/HDAC-mediated transcriptional repression is important for maintaining proliferative potential in a wide range of cell types.Citation12 In accordance, studies on Ski in connection with members of the Sin3/HDAC complex have linked its function to regulation of cell proliferation and differentiation.
Ski and the NuRD/HDAC Repressor Complex
To date, it was believed that Ski is exclusively active in Sin3/HDAC or N-CoR/HDAC complexes when functioning as a transcriptional repressor. Only recently, however, we discovered that, in the cortical plate, Ski forms complexes with MTA2, a component of the NuRD/HDAC complex.Citation13 For this, we performed immunoprecipitation experiments using cortical lysates, and applied an antibody-based proximity ligation assay, which allowed us to visualize MTA2-Ski complex formation in situ in the cortical plate of mouse embryos. Using similar methods, this approach led to the detection of a previously unidentified Ski-interacting protein, the chromatin-remodeling factor Satb2, which is expressed in callosal neurons during cortical development. Satb2 and its family member Satb1 have been shown to regulate gene expression by binding matrix attachment regions (MARs) and promoting higher-order chromatin organization at target sequences.Citation14 In the developing telencephalon, Satb2 binds to multiple MAR sequences within its target gene Ctip2, and represses activation of Ctip2 transcription in callosal neurons. Notably, Ctip2 is involved in the specification of deep layer neurons that project to subcortical targets, whereas Satb2 specifies the identity of callosal projecting neurons. We found that in Ski-deficient mice, Satb2-expressing callosal neurons ectopically expressed Ctip2, eventually causing them to change their identity and project ectopically to subcortical targets. Our experiments provided evidence that interactions of Satb2 and MTA2 with MAR sequences are independent of Ski, and that Ski is essential for recruiting HDAC1 to the protein assembly. Thus, Ski enables the Satb2-containing protein assembly to act as a functional NuRD/HDAC inhibitory complex and to promote Ctip2 transcriptional repression in callosal neurons. Studies with mice mutant for NuRD components have indicated that this co-repressor complex is largely involved in cell type specification.Citation12 Hence, this raises the question of whether Ski/NuRD/HDAC complexes are usually involved in regulating cell fates, whereas Ski/Sin3/HDAC protein assemblies control cell proliferation and differentiation.
Diverse Functions of Ski During Cortical Development
During embryonic development, proliferating neural stem and progenitor cells located in the germinal zones of the dorsal telencephalon give rise to molecularly and functionally distinct projection neurons of the neocortex. Initially, neuroepithelial/radial glial cells in the ventricular zone (VZ) of the dorsal forebrain, referred to as apical progenitor cells, generate the earliest born neurons. Later in development, apical progenitors additionally produce the so-called basal or intermediate progenitor cells that constitute the adjacent telencephalon-specific subventricular zone (SVZ).Citation15 From mouse embryonic day (E) 11.5 to E17.5, the heterogeneous pool of progenitor cells in the VZ and SVZ give rise to projection neurons, which dependent on their birthdates, are targeted to their respective laminar positions in the cortical plate. Early-born neurons populate the deep layers VI and V, whereas late-generated neurons migrate past the early-born neurons and constitute the upper layers IV, III and II of the neocortex.Citation16
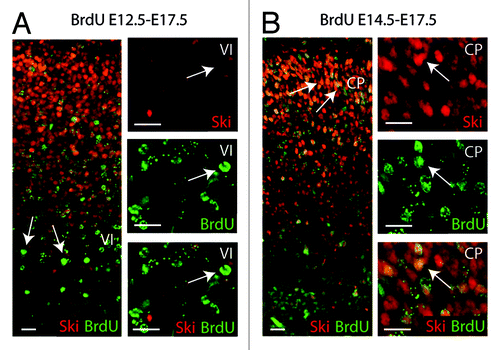
We have recently determined the spatial and temporal distribution of the Ski protein in the developing dorsal telencephalon.Citation13 Our detailed analysis revealed that Ski is not ubiquitously expressed but is selectively present in proliferating progenitor cells in the VZ and in subtypes of differentiated projection neurons of the cortical plate. Interestingly, the apical progenitor cells expressed Ski at high levels throughout development,Citation13 while its expression in projection neurons was transient and particularly high in young neurons (). Using Ski−/− mice, we found that, as a result of Ski deletion, cells differentiated precociously into neurons, leading to a reduced progenitor pool at initial stages of neural development. How does Ski prevent proliferating apical progenitors from premature cell cycle exit and precocious initiation of neurogenesis? The best-characterized function of Ski, which was assessed under overexpression conditions and in cancer cells, is to negatively regulate transforming growth factor β (TGFβ) signaling via interaction with Smad proteins.Citation11 Binding of TGFβ to its receptors results in phosphorylation of the receptor-regulated Smads, which then translocate into the nucleus where they bind to DNA and activate the transcription of target genes. Ski was shown to repress the transcriptional activation through its concurrent interaction with Smads and members of the Sin3/HDAC complex.Citation17-Citation21 Notably, TGFβ and its cellular mediators are expressed in the developing mouse forebrain. Moreover, studies using primary cultures of cortical cells revealed antimitotic effects of TGFβ on progenitor cells and increased expression of neuronal markers in TGFβ-treated cortical neurons.Citation22 Thus, it is conceivable that Ski, as a repressor of the TGFβ pathway, modulates its action during cortical development through recruitment of the Sin3/HDAC complex to Smads, thereby fine-tuning the balance between proliferation and differentiation of progenitor cells.
Figure 1. Ski is expressed in young cortical neurons. Distributions of bromodeoxyuridine (BrdU) birth date-labeled cells in E17.5 neocortex after BrdU injection at E12.5 (A) and at E14.5 (B) are shown. E12.5-born neurons located in layer VI are only weakly positive for Ski (arrows in A and in higher magnifications). E14.5-born neurons located in superficial layers of the cortical plate (CP) are strongly positive for Ski (arrows in B and in higher magnifications). Scale bars represent 20 µm.
In contrast, we found that Ski expression in postmitotic cells of the developing telencephalon ensures proper cell specification. Loss of Ski triggered a change in identity of callosal neurons by prompting axons to descend along the corticospinal tract instead of extending across the corpus callosum.Citation13 In this case, Ski associated with the NuRD/HDAC complex and the cell-type specific chromatin-remodeling factor Satb2 to repress target genes such as Ctip2. Based on in situ hybridization assays, we found that, besides Ctip2, the expression of other deep layer-specific genes was misregulated. They displayed increased expression levels in deep layers and an expansion of expression into upper layers upon loss of Ski. This finding suggested that Ski not only specifies callosal neurons, but might also be essential for the specification of subcerebrally projecting neurons. Notably, subpopulations of deep-layer neurons express Satb1, a family member of Satb2, which also interacts with Ski (unpublished data), and is likely also to be involved in cell specification processes.
In summary, based on a large set of in vitro data and our recently obtained findings in vivo in cortical development, the variable duration of Ski expression in proliferating and postmitotic cells might reflect the differential temporal requirements for particular genetic programs to be in place in these paradigms. It is conceivable that high and persistent expression of Ski is required for regulating long-lasting, continuous processes such as cell proliferation and differentiation as observed under overexpression conditions, in cancer cells, and in cortical progenitor cells during embryonic development.
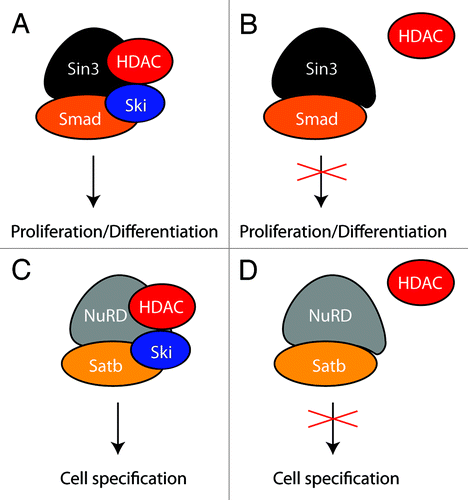
In contrast, transient Ski expression would be sufficient to impose particular, temporally restricted genetic programs to initiate proper cell specification as observed for callosal projection neurons. Finally, based on the currently available data, Ski appears to regulate these different processes, at least in part, by recruiting HDAC to distinct co-repressor complexes ().
Figure 2. A mechanistic model for the repressor function of Ski. Ski is required to assemble a functional Sin3 (A) and NuRD (C) repressor complex at target sequences. In the absence of Ski, recruitment of HDAC to the Sin3 (B) and NuRD (D) complex is impaired resulting in misregulation of cell proliferation/differentiation (A,B) and cell specification (C,D), respectively.
| Abbreviations: | ||
| HDAC | = | histone deacetylase |
| NuRD | = | nucleosome remodeling and deacetylase repressor |
| N-coR | = | nuclear receptor co-repressor |
| SMRT | = | silencing mediator for retinoid and thyroid hormone receptors |
| MAR | = | matrix attachment region |
| VZ | = | ventricular zone |
| SVZ | = | subventricular zone |
| TGFβ | = | transforming growth factor beta |
Acknowledgments
We thank Ned Mantei for valuable comments on the manuscript. This work was supported by the Swiss National Science Foundation, the Novartis Foundation, Swiss Life, and the Botnar Foundation.
References
- Nagase T, Mizuguchi G, Nomura N, Ishizaki R, Ueno Y, Ishii S. Requirement of protein co-factor for the DNA-binding function of the human ski proto-oncogene product. Nucleic Acids Res 1990; 18:337 - 43; http://dx.doi.org/10.1093/nar/18.2.337; PMID: 2183181
- Wilson JJ, Malakhova M, Zhang R, Joachimiak A, Hegde RS. Crystal structure of the dachshund homology domain of human SKI. Structure 2004; 12:785 - 92; http://dx.doi.org/10.1016/j.str.2004.02.035; PMID: 15130471
- Wu JW, Krawitz AR, Chai J, Li W, Zhang F, Luo K, et al. Structural mechanism of Smad4 recognition by the nuclear oncoprotein Ski: insights on Ski-mediated repression of TGF-beta signaling. Cell 2002; 111:357 - 67; http://dx.doi.org/10.1016/S0092-8674(02)01006-1; PMID: 12419246
- Sleeman JP, Laskey RA. Xenopus c-ski contains a novel coiled-coil protein domain, and is maternally expressed during development. Oncogene 1993; 8:67 - 77; PMID: 8423997
- Amaravadi LS, Neff AW, Sleeman JP, Smith RC. Autonomous neural axis formation by ectopic expression of the protooncogene c-ski. Dev Biol 1997; 192:392 - 404; http://dx.doi.org/10.1006/dbio.1997.8780; PMID: 9441676
- Kaufman CD, Martínez-Rodriguez G, Hackett PB Jr.. Ectopic expression of c-ski disrupts gastrulation and neural patterning in zebrafish. Mech Dev 2000; 95:147 - 62; http://dx.doi.org/10.1016/S0925-4773(00)00351-8; PMID: 10906458
- Nomura T, Khan MM, Kaul SC, Dong HD, Wadhwa R, Colmenares C, et al. Ski is a component of the histone deacetylase complex required for transcriptional repression by Mad and thyroid hormone receptor. Genes Dev 1999; 13:412 - 23; http://dx.doi.org/10.1101/gad.13.4.412; PMID: 10049357
- Zhao HL, Ueki N, Marcelain K, Hayman MJ. The Ski protein can inhibit ligand induced RARalpha and HDAC3 degradation in the retinoic acid signaling pathway. Biochem Biophys Res Commun 2009; 383:119 - 24; http://dx.doi.org/10.1016/j.bbrc.2009.03.141; PMID: 19341714
- Hayakawa T, Nakayama J. Physiological roles of class I HDAC complex and histone demethylase. J Biomed Biotechnol 2011; 2011:129383; http://dx.doi.org/10.1155/2011/129383; PMID: 21049000
- Kokura K, Kaul SC, Wadhwa R, Nomura T, Khan MM, Shinagawa T, et al. The Ski protein family is required for MeCP2-mediated transcriptional repression. J Biol Chem 2001; 276:34115 - 21; http://dx.doi.org/10.1074/jbc.M105747200; PMID: 11441023
- Bonnon C, Atanasoski S. c-Ski in health and disease. Cell Tissue Res 2012; 347:51 - 64; http://dx.doi.org/10.1007/s00441-011-1180-z; PMID: 21647564
- McDonel P, Costello I, Hendrich B. Keeping things quiet: roles of NuRD and Sin3 co-repressor complexes during mammalian development. Int J Biochem Cell Biol 2009; 41:108 - 16; http://dx.doi.org/10.1016/j.biocel.2008.07.022; PMID: 18775506
- Baranek C, Dittrich M, Parthasarathy S, Bonnon CG, Britanova O, Lanshakov D, et al. Protooncogene Ski cooperates with the chromatin-remodeling factor Satb2 in specifying callosal neurons. Proc Natl Acad Sci U S A 2012; 109:3546 - 51; http://dx.doi.org/10.1073/pnas.1108718109; PMID: 22334647
- Britanova O, Akopov S, Lukyanov S, Gruss P, Tarabykin V. Novel transcription factor Satb2 interacts with matrix attachment region DNA elements in a tissue-specific manner and demonstrates cell-type-dependent expression in the developing mouse CNS. Eur J Neurosci 2005; 21:658 - 68; http://dx.doi.org/10.1111/j.1460-9568.2005.03897.x; PMID: 15733084
- Merkle FT, Alvarez-Buylla A. Neural stem cells in mammalian development. Curr Opin Cell Biol 2006; 18:704 - 9; http://dx.doi.org/10.1016/j.ceb.2006.09.008; PMID: 17046226
- Leone DP, Srinivasan K, Chen B, Alcamo E, McConnell SK. The determination of projection neuron identity in the developing cerebral cortex. Curr Opin Neurobiol 2008; 18:28 - 35; http://dx.doi.org/10.1016/j.conb.2008.05.006; PMID: 18508260
- Akiyoshi S, Inoue H, Hanai J, Kusanagi K, Nemoto N, Miyazono K, et al. c-Ski acts as a transcriptional co-repressor in transforming growth factor-beta signaling through interaction with smads. J Biol Chem 1999; 274:35269 - 77; http://dx.doi.org/10.1074/jbc.274.49.35269; PMID: 10575014
- Luo K, Stroschein SL, Wang W, Chen D, Martens E, Zhou S, et al. The Ski oncoprotein interacts with the Smad proteins to repress TGFbeta signaling. Genes Dev 1999; 13:2196 - 206; http://dx.doi.org/10.1101/gad.13.17.2196; PMID: 10485843
- Sun Y, Liu X, Eaton EN, Lane WS, Lodish HF, Weinberg RA. Interaction of the Ski oncoprotein with Smad3 regulates TGF-beta signaling. Mol Cell 1999; 4:499 - 509; http://dx.doi.org/10.1016/S1097-2765(00)80201-4; PMID: 10549282
- Xu W, Angelis K, Danielpour D, Haddad MM, Bischof O, Campisi J, et al. Ski acts as a co-repressor with Smad2 and Smad3 to regulate the response to type beta transforming growth factor. Proc Natl Acad Sci U S A 2000; 97:5924 - 9; http://dx.doi.org/10.1073/pnas.090097797; PMID: 10811875
- Ueki N, Hayman MJ. Direct interaction of Ski with either Smad3 or Smad4 is necessary and sufficient for Ski-mediated repression of transforming growth factor-beta signaling. J Biol Chem 2003; 278:32489 - 92; http://dx.doi.org/10.1074/jbc.C300276200; PMID: 12857746
- Vogel T, Ahrens S, Büttner N, Krieglstein K. Transforming growth factor beta promotes neuronal cell fate of mouse cortical and hippocampal progenitors in vitro and in vivo: identification of Nedd9 as an essential signaling component. Cereb Cortex 2010; 20:661 - 71; http://dx.doi.org/10.1093/cercor/bhp134; PMID: 19587023