Abstract
Long interspersed nuclear element 1 (LINE-1) retrotransposons are mutagens that are capable of generating deleterious mutations by inserting themselves into genes and affecting gene function in the human genome. In normal cells, the activity of LINE-1 retrotransposon is mostly repressed, maintaining a stable genome structure. In contrast, cancer cells are characterized by aberrant expression of LINE-1 retrotransposons, which, in principle, have the potential to contribute to genomic instability. The mechanistic pathways that regulate LINE-1 expression remain unclear. Using deep-sequencing small RNA analysis, we identified a subset of differentially expressed endo-siRNAs that directly regulate LINE-1 expression. Detailed analyses suggest that these endo-siRNAs are significantly depleted in human breast cancer cells compared with normal breast cells. The overexpression of these endo-siRNAs in cancer cells markedly silences endogenous LINE-1 expression through increased DNA methylation of the LINE-1 5′-UTR promoter. The finding that endo-siRNAs can silence LINE-1 activity through DNA methylation suggests that a functional link exists between the expression of endo-siRNAs and LINE-1 retrotransposons in human cells.
Introduction
Long interspersed nuclear element-1 (LINE-1 or L1) retrotransposons are the most prolific class of retrotransposable elements, comprising 17% of human genomic sequences. There are ~6,000 full-length LINE-1 elements in the human genome, at least 100 of which are classified as active elements or retrotransposition competents.Citation1,Citation2 An active LINE-1 element is ~6 kb long and is composed of the 5′-untranslated region (5′-UTR), which harbors an internal promoter, two open reading frames (ORF1 and ORF2), and the 3′-UTR, which includes a poly-A tail.Citation3 ORF1 encodes a p40 protein with RNA-binding and chaperone activities,Citation4 whereas ORF2 encodes a protein of 150 kDa with endonuclease and reverse transcriptase activities.Citation5,Citation6 Both ORF proteins are required for autonomous LINE-1 retrotransposition activity.Citation7
LINE-1 is an insertional mutagen that is capable of altering the genome through disrupting genes, altering splicing sites, increasing the frequency of recombination, and negatively affecting the stability of the genome because of its ability to create breaks in genomic DNA breaks during the process of LINE-1 mobilization or retrotransposition (reviewed in refs. Citation8 and Citation9). In addition, LINE-1 mediates the retrotransposition of nonautonomous SINE elements such as Alus and retrogenes,Citation10,Citation11 thereby altering the genomic structure in myriad ways. There are at least 60 known human disease-causing insertions of LINE-1s, Alus and SVAs.Citation12 Although knowledge about LINE-1 retrotransposition in somatic tissues is still scarce, increasing evidence suggests that LINE-1 is expressed in some somatic cellsCitation13 and that ongoing LINE-1 retrotransposition causes somatic mosaicism in neuronal progenitor cells and during embryogenesis.Citation13,Citation14 A recent study surveying lung tumors and comparing their genomes against normal tissues suggested that tumors exhibit high frequencies of LINE-1 retrotransposition events that are not present in the adjacent normal tissues.Citation15 Because of the potential harmful impact of LINE-1 elements on genome integrity, LINE-1 expression is believed to be held in check through a variety of genome defense mechanismsCitation16,Citation17 and is mostly undetectable in normal cells. In contrast, the majority of human cancers, including breast cancer,Citation18 exhibit high levels of LINE-1 expression, which can be one of the driving forces causing an accelerated rate of mutations and genomic instability. These defects in LINE-1 expression have been attributed to transcription activation associated with the DNA methylation of LINE-1 promoters.Citation19,Citation20 It was recently reported that the hypomethylation of LINE-1 promoters activates an alternate promoter of the MET oncogene in cancer cells.Citation21
Several studies performed in the DrosophilaCitation22,Citation23and mouse germlinesCitation24 have now suggested that naturally occurring endogenous siRNAs (endo-siRNAs), possibly derived from retrotransposons or from complementary annealed transcripts, can repress LINE-1 expression through an RNAi pathway, acting as a constant genome defense system. However, whether such endo-siRNAs exist in human somatic cells remains unclear, and thus endo-siRNAs are not well characterized. One piece of evidence that supports a role for natural endo-siRNAs is the detection of 21-nt siRNAs in cultured human cells using strand-specific RNA probes for the LINE-1 5′-UTR promoter.Citation16 However, whether these hybridization signals represent a pool of an siRNA population or functional siRNA duplexes remains unknown. Although bona fide endo-siRNAs have yet to be isolated, we and others have shown that the knockdown of components of the RNAi machinery, such as Dicer1,Citation16,Citation25 increases the rate of human LINE-1 retrotransposition activity, suggesting that a possible link exists between small RNAs and the epigenetic silencing of retrotransposons. Despite extensive analyses and their significant role, little is known about small RNAs or their potential function as regulators of LINE-1 elements in the human genome.
Endo-siRNAs are small non-coding RNAs of 20–24 nt that control target gene expression through their secondary structure and by recruiting chromatin-targeted RNAi silencing components to form transcriptionally silent heterochromatin structures.Citation26,Citation27 Using high-throughput deep sequencing, we recently identified a subset of repeat-associated endo-siRNAs that are differentially expressed between normal breast cells and breast cancer cells. Interestingly, the identified endo-siRNAs are perfectly complementary in sequence to the LINE-1 bidirectional promoters. In this study, we characterized the functions of these differentially expressed endo-siRNAs in breast cancer cells. Here, we demonstrate that the overexpression of the endo-siRNAs in cancer cells resulted in marked silencing of LINE-1 expression through global hypermethylation of the LINE-1 promoter. The finding that endo-siRNAs can repress LINE-1 activity through DNA methylation provides new insight into the function of endo-siRNA in the silencing of retrotransposons and the maintenance of genome integrity in human cells.
Results
Aberrant LINE-1 expression in breast cancer cells
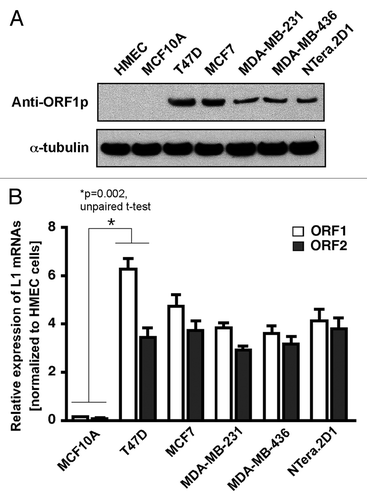
Earlier studies revealed that the levels of LINE-1 transcripts are significantly elevated in human breast cancers and cancer-derived cell lines.Citation28,Citation29 A recent study of clinical specimens of breast cancers has also shown that the expression of the LINE-1 ORF1 protein is widespread in a range of breast tumors,Citation18 but the mechanisms that influence LINE-1 expression remain unknown. To gain insight into the mechanisms by which LINE-1 contributes to breast cancers, we first determined the expression level of LINE-1-encoded ORF1 protein in normal breast epithelial cells (HMEC and MCF10A) and breast cancer cells (T47D, MCF-7, MDA-MB-231 and MDA-MB-436) by western blotting, as described previously.Citation18 Human embryonic carcinoma NTera.2D1 cells, which are known to express high levels of LINE-1 mRNA and the encoded proteins as a results of altered transcription start sites,Citation30,Citation31 were used as a positive control. As shown in , the LINE-1 ORF1 protein was significantly overexpressed in all breast cancer cells but not in non-tumorigenic MCF10A breast cells which were derived from primary mammary epithelial HMEC tissue. To further confirm this finding, the relative expression levels of LINE-1 mRNA were analyzed using real-time quantitative RT-PCR. These assays showed that both LINE-1-encoded ORF1 and ORF2 mRNAs were significantly overexpressed in all breast cancer cells (). Notably, the relative expression of LINE-1 mRNA and the encoded ORF1 protein was markedly higher in non-invasive T47D cancer cells compared with moderate (MCF7)-to-invasive (MDA-MB-231) cancer cells. The findings that LINE-1 is overexpressed in non-invasive cancer cells, suggest that LINE-1 expression in principle has the potential to contribute to genomic instability in these cells.
Figure 1. Aberrant expression of LINE-1 retrotransposons in breast cancer cells. (A) The LINE-1-encoded ORF1 protein was detected by western blotting of whole-cell lysates from normal and breast cancer cells. NTera.2D1 human embryonic carcinoma cells were used as positive controls. For protein normalization, mouse α-tubulin was used as a loading control. (B) The LINE-1 transcripts (L1 mRNAs) derived from the LINE-1 ORFs were detected via qRT-PCR with primers specific for the LINE-1 ORF1 and ORF2 sequences. The data are shown as the fold change compared with the HMECs after normalization to the HPRT1 housekeeping gene. Each point represents the average from four independent experiments. Unpaired t-test, p = 0.002. Error bars indicate s.d. (n = 4).
Breast cancer cells support in vivo LINE-1 retrotransposition activity
To investigate whether breast cancer cells can also support LINE-1 retrotransposition activity, we transfected the engineered human LINE-1 retrotransposition cassette into normal MCF10A cells and breast cancer cell lines (T47D, MCF-7, MDA-MB-231 and MDA-MB-436) to create a cell-based assay that accurately detects endogenous LINE-1 retrotransposition events. In this assay, human embryonic carcinoma NTera.2D1 cells were used as a positive control. The retrotransposition cassette (pRP99-L1-EGFP) contained a full-length human L1RP retrotransposon (driven by its native promoter) tagged at its 3′UTR with an antisense EGFP expression cassette.Citation7 The EGFP gene was disrupted by a 960 bp sequence of the γ–globin intron in the same orientation as the LINE-1 transcript (). This arrangement ensures that functional EGFP expression occurs only after an LINE-1 retrotransposition event or insertional mutagenesis, that is, following LINE-1 expression, γ–globin intron splicing, reverse transcription, and insertion of a copy of LINE-1 into the genomic DNA of the host cell. The levels of EGFP expression in the pRP99-L1-EGFP transfected cells could be measured under UV light, allowing us to detect near real-time LINE-1 retrotransposition events in living cells without cell staining. An inactive form of LINE-1 (pRP99-ΔL1-EGFP) containing missense mutations in ORF1 to abrogate retrotransposition activity was used as a negative control.Citation7 In the resulting assay, EGFP-positive cells were not detected in any of the normal MCF10A-transfected cells, even after several passages (). In contrast, all breast cancer cells exhibited retrotransposition events, averaging 687 to 1,063 events per 100,000 cells, while the mutant pRP99-ΔL1-EGFP showed no retrotransposition (not shown). Interestingly, the relative LINE-1 retrotransposition frequency was significantly higher in non-invasive T47D cells (1,063 events on average), compared with MCF7 (890 events), MDA-MB-231 (717 events), MDA-MB-436 (687) and NTera.2D1 (775) cells, suggesting that T47D cells support high levels of LINE-1 retrotransposition activity, which is consistent with the high levels of LINE-1 mRNA and the encoded ORF1 protein observed in these cells.
Figure 2. The LINE-1 retrotransposition cassette. (A) Schematic diagrams of the pRP99-L1-EGFP expression cassettes used for LINE-1 retrotransposition assays. LINE-1 transcription is driven by its own 5′UTR, which harbors an internal promoter. This LINE-1 retrotransposon contains an intron-interrupted EGFP reporter in the 3′UTR region with its own CMV promoter and polyadenylation signal. The EGFP indicator cassette is in an antisense orientation relative to LINE-1. Only when EGFP is transcribed from the LINE-1 promoter, spliced, reverse transcribed and integrated into the genome does a cell become EGFP positive. Arrows depict the location of the geno-5 (left) and geno-3 (right) primers used in the PCR assay shown below. SD, splice donor; SA, splice acceptor. (B) Detection of sustained retrotransposition events in breast cancer cells. The number of EGFP-positive cells was plotted over time for pRP99-L1-EGFP transfected cells. Each time point represents cell populations from three independent experiments. Sampling and analysis were performed at 1, 3, 5, 8, 12, and 15 d after the plating of transfected cells. The X-axis indicates the number of EGFP-positive cells per 105 cells analyzed. The error bars indicate s.d (C) PCR analysis of retrotransposed cells. The geno-5 and geno-3 primers, flanking the intron in EGFP, were used for PCR amplification of genomic DNA, and the obtained products were analyzed on a 1.2% agarose gel. PCR products of 1.49 kb (corresponding to the intron-containing vector) and 530 bp (corresponding to the retrotransposed insertion that lacks the 909 bp intron) are shown. As a negative control, genomic DNA from the parental cells (P) was used. The symbols + and – represent the active LINE-1 (pRP99-L1-EGFP) and the inactive LINE-1 (pRP-ΔL1-EGFP) transfected cells, respectively. Vector, 1 ng plasmid DNA; Marker, 1 kb-plus DNA marker.
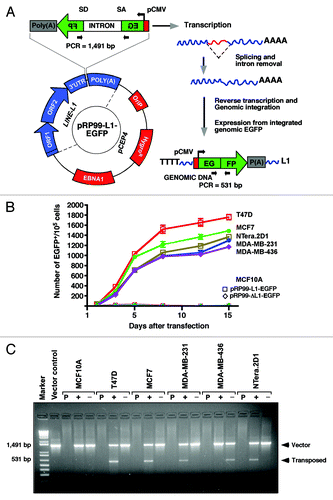
To determine whether the observed EGFP expression was a result of in vivo LINE-1 retrotransposition activity, we conducted a PCR based genomic DNA analysis to confirm the presence of retrotransposed or spliced EGFP in these cells. Intron-less EGFP (~0.5 kb in size) was amplified from all cancer cells transfected with the active pRP99-L1-EGFP cassette (shown with a plus symbol) but not from cells transfected with the inactive pRP99-ΔL1-EGFP (shown with a minus symbol), indicating the occurrence of retrotransposition events (). These observations suggest that de novo LINE-1 retrotransposition indeed occurs in breast cancer cells but not in normal MCF10A breast cells. In principle, MCF10A cells might present genomic defense mechanisms that are responsible for silencing endogenous LINE-1 expression.
Depletion of Dicer1 or Ago2 activates LINE-1 expression
LINE-1 is expressed at very low levels in somatic cells, if at all. In contrast, the overexpression of LINE-1 is a characteristic feature of many cancer-derived cells and germlines,Citation32,Citation33 as well as a variety of transformed cell lines.Citation13 The mechanisms that activate LINE-1 expression in these cells are currently unknown. Recent discoveries have revealed the existence of Piwi-interacting RNAs (piRNAs) which silence retrotransposon expression in the germline of Drosophila and the mouse.Citation34 In addition, small RNA sequencing from mouse oocytesCitation35 revealed that some siRNAs derived from retrotransposons silence LINE-1 elements in trans, suggesting that the multiple mechanisms may control LINE-1 expressions in mammals. Studies on Drosophila and S. pombe suggest that endo-siRNAs, in complex with Ago2 proteins, can identify their transposon targets through sequence-specific recognition, thus enabling the targeting of chromatin-modifying enzymes to the repeat sequences that they modify.Citation34,Citation36 Consistent with this finding, a mutation in the Ago2 protein in plants and fission yeasts is correlated with loss of siRNA-directed transcriptional silencing.Citation37 A recent report on Drosophila indicates that naturally occurring some endo-siRNAs silence transposon expression by targeting their promoters at the transcriptional and post-transcriptional levels.Citation27,Citation38 However, the few endo-siRNAs that have been identified thus far originated from mouse, Drosophila, Arabidopsis, and C. elegans. Direct evidence for the existence of such endo-siRNAs in the human genome is still lacking.
The human LINE-1 5′-UTR promoter is known to contain both the sense and anti-sense promoters,Citation39 which can potentially lead to the production of dsRNAs, which are in turn, processed by Dicer1 to yield a series of LINE-1-specific siRNAs.Citation16 A previous study by our group on chicken-human hybrid DT40 cells showed that there was a reduced accumulation of small RNAs overall in Dicer-deficient cells and thus, increased expression of LINE-1 transcripts.Citation25 Although these studies suggested that the RNAi pathway is required for the silencing of the LINE-1 elements, we obtained no direct evidence for the existence of small RNAs that can control human LINE-1 retrotransposons. Endo-siRNAs are small non-coding RNAs that arise from convergent transcription units or from structured genomic loci through the action of Dicer1. These small RNAs bind to Ago2 protein complexes and repress retrotransposons in somatic cells of Drosophila.Citation40 In humans, the existence of such mechanisms remains unclear.
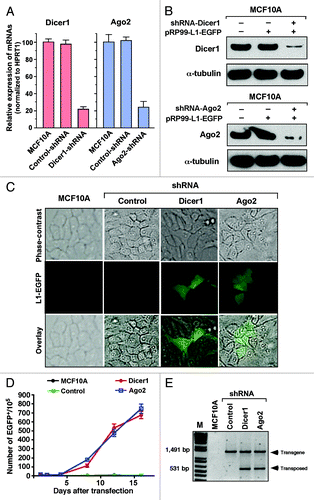
To test whether the RNAi pathway is required for the production and association with Ago2 proteins to silence human LINE-1 elements, we used recently published Dicer1 and Ago2-specific shRNA sequence separately in normal MCF10A breast cells.Citation41 An shRNA targeting the luciferase gene was used as a negative control. Using qRT-PCR analysis, we achieved knockdown efficiencies of 78 ± 3.5% and 76 ± 7.7% for the Dicer1 and Ago2 genes, respectively (). These results were further confirmed by western blotting against Dicer1 and Ago2 antibodies (). The effects of Dicer1 and Ago2 knockdown on LINE-1 expression were assessed by transfecting MCF10A cells with the active LINE-1 pRP99-L1-EGFP retrotransposition cassette. LINE-1-retrotransposed EGFP-positive cells were readily detected in Dicer1 and Ago2 knockdown cells within 72 h post-transfection but not in any of the parental or control shRNA-transfected cells (). Using flow cytometric analysis, we compared the LINE-1 retrotransposition frequencies in these cells. Both the Dicer1- and Ago2-knockdown cells showed high levels of LINE-1 retrotransposition activity, averaging 225 and 240 events for Dicer1 and Ago2, respectively, whereas cells transfected with control shRNA and parental MCF10A cells did not express any EGFP for at least 16 d (). To further validate these findings, we performed a PCR-based genomic DNA analysis to confirm the presence of retrotransposed LINE-1 or spliced EGFP in these cells (). Transposed LINE-1 (~0.5 kb in size) was indeed amplified from MCF10A cells transfected with the Dicer1 and Ago2 shRNAs but not from cells transfected with the control shRNA, indicating the occurrence of LINE-1 retrotransposition events upon depletion of Dicer1 and Ago2 proteins. These observations indicate that there is a link between small RNAs processed by Dicer1/Ago2 and the silencing of LINE-1 expression. Taken together, these results suggest that some naturally occurring small RNAs in normal MCF10A breast cells specifically target LINE-1 expression in association with components of the RNAi machinery, such as Dicer1 and Ago2 protein complexes, akin to endo-siRNAs in Drosophila somatic cells.
Figure 3. Depletion of Dicer1 and Ago2 activates LINE-1 expression. (A) Quantitative real-time RT-PCR analysis of endogenous Dicer1 and Ago2 mRNAs in normal MCF10A cells that were stably transfected with shRNAs targeting the Dicer1 and Ago2 genes. An shRNA targeting the luciferase gene was used as a negative control. The data are shown as the relative fold changes of Dicer1 and Ago2 mRNAs with respect to the control HPRT1. (B) western blot of endogenous Dicer1 and Ago2 proteins knocked down in MCF10A cells. α-tubulin was used as an internal control. (C) Dicer1- and Ago2-depleted MCF10A cells were transiently transfected with the pRP99-L1-EGFP expression cassette. The analyses of the parental MCF10A cells and cells transfected with control shRNA were performed in parallel. EGFP-expressing cells were readily detected 3 d after transfection. The EGFP signal was digitally overlaid on the corresponding phase-contrast image. (D) The time course of LINE-1 retrotransposition events in Dicer1- and Ago2-knockdown cells. Number of gated EGFP-positive cells recorded at various time points. Each time point represents cell populations from three independent transfections. The error bars indicate s.d. (n = 3). (E) Confirmation of retrotransposition events using PCR, as revealed by a 531 bp band observed in MCF10A cells in which the Dicer1 and Ago2 genes were depleted, but not in cells treated with control shRNA or parental MCF10A cells. The 1,491 bp band represents the intron-containing pRP99-L1-EGFP vector.
Identification and validation of endo-siRNA function
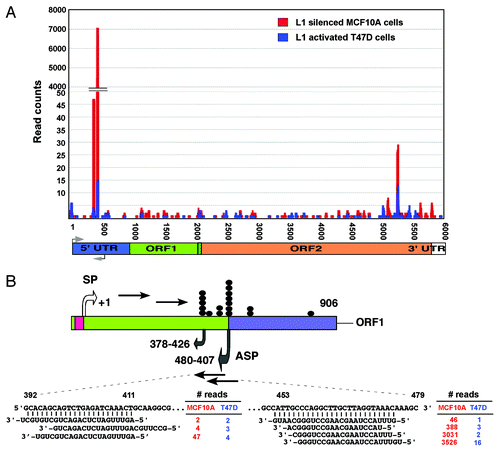
To identify potential endogenous small RNAs in the human genome, we recently sequenced two small RNA libraries from normal MCF10A breast cells, which show little or no LINE-1 expression, and from T47D breast cancer cells, which strongly overexpress LINE-1 elements, using the Illumina genome analyzer (manuscript in preparation). Following the normalization and subsequent bioinformatics analysis, we identified two subsets of small RNAs that were differentially expressed between the MCF10A and T47D cells (data not shown). These antisense small RNAs were matched perfectly complementary to the 5′-UTR of the most active Homo sapiens-specific LINE-1Hs subfamilies (). Annotation of the small RNAs using the deepBase mapping databaseCitation42 (version GRCh37, UCSC hg19) revealed that these species represented repeat-associated endo-siRNAs and mapped mainly to the deepBase sequences hgur000097447 and hgur000380276 (termed endo392 and endo453, respectively). A comparison with the position of the LINE-1 5′-UTR revealed that enrichment of these endo-siRNAs yields two distinct peaks at positions of 392 to 479 nucleotides of the LINE-1 element corresponding to the bidirectional promoters of the LINE-1 5′-UTR. Interestingly, a previous study found that the deletion of this region of the LINE-1 5′-UTR increased the expression of LINE-1 elements in cultured human cells,Citation16 indicating that this region may be the target site for small RNA binding. The expression profiles of the endo-siRNAs and their read counts were distinctly different between the normal MCF10A breast cells and T47D breast cancer cells (). The reason for this differential expression between normal and cancer cells is currently unknown. The high levels of expression of the homogeneous population of endo-siRNAs, with the majority of species exhibiting lengths of 20 to 22 nt, raises the possibility that these endo-siRNAs are likely to play a key role in silencing human LINE-1 elements.
Figure 4. Comparison of LINE-1-specific endo-siRNA abundance in MCF10A and T47D cells. (A) Read counts of endo-siRNAs and their binding sites on LINE-1 (L1) retrotransposons are shown. (B) Structure of the LINE-1Hs 5′-UTR showing the sense (SP) and antisense (ASP) promoters within the ~906 nucleotides of the promoter. Transcription start sites are given, numbered according to the sense strand. Known transcription start sites of ASP involved in the regulation of LINE-1 are denoted by closed dot symbols. Grey arrows indicate the bidirectional transcripts of the LINE-1 5′-UTR. Sequences of two differentially expressed endo-siRNAs, read counts, and their binding positions on the LINE-1 5′–UTR sequences are shown.
To investigate whether the identified endo-siRNAs are also present in other types of breast cancer cells, we performed northern blot analysis using low-MW RNA isolated from various breast cancer cells and probed with DNA oligos complementary to the endo-siRNA sequences hgur000097447 (endo392) and hgur000380276 (endo453). As a loading control, hsa-miR16 was used (). The results showed that compared with normal breast HMECs or MCF10A cells, the endo-siRNAs were weakly detected in all breast cancer cells. To accurately quantify the expression levels of endo-siRNAs, we used a custom-designed TaqMan assay that employs a stem-looped primer for reverse transcription and a sequence-specific MGB probe. The fold changes in breast cancer cells vs. normal cells were analyzed using real-time qRT-PCR (). In this assay, SnoRNA37A served as a normalizing control. We found that both of the endo-siRNAs were significantly underexpressed in all breast cancer cells, which is consistent with the high levels of LINE-1 mRNA and the encoded ORF1 protein observed in these cells. Strikingly, both of these endo-siRNAs were found to exhibit significant secondary RNA structures (Mfold prediction) with calculated free energies (∆G037 = –30.2 and ∆G037 = –30.7 for endo453 and endo392, respectively) that may be required for their interaction with Argonaute proteins.Citation43 Moreover, using the UCSC genome browser, we found that these endo-siRNA sequences did not show any significant homology to any mRNAs or RefSeq cDNAs by querying the NCBI non-redundant database (data not shown), indicating that the expression of endo-siRNAs may specifically regulate the expression levels of human LINE-1 elements.
Figure 5. Endo-siRNA mediates LINE-1 silencing. (A) northern blot analysis of endo-siRNAs. Low MW total RNAs were probed with the sense strand of endo-siRNA sequences, and the resulting signals were detected after exposure to X-film for 3 d. As a small RNA loading control, hsa-miR-16 was used. Labeled Decade RNA markers were used as size markers. (B) Differential expression of endo-siRNAs in various types of breast cancer cells determined using real-time qRT-PCR. The fold changes were determined by comparing the ∆CT value of each endo-siRNA after normalization to the control Sno37A small RNA. The data are shown as the ratio of the mean signal in cancer cells/mean signal in normal cells. A negative value indicates reduced expression of endo-siRNAs in cancer cells compared with MCF10A cells. (C) A schematic of the construct used for the dual luciferase report assay is shown in the top panel. The synthetic siRNAs used in this study are shown at the bottom. Luciferase reporter assay detecting expression from in HEK293T cells transfected with scrambled, endo453 and endo392 siRNAs. Cells transfected with firefly luciferase (under control of the LINE-1Hs 5′-UTR) and the Renilla luciferase reporter plasmid without any siRNA sequences were used as a negative control. The firefly luciferase activity was normalized to the Renilla luciferase activity. The data are shown as the relative luciferase activity of endo-siRNA-treated cells with respect to the control cells. Unpaired t-test, p = 0.005. Error bars indicate s.d. (n = 9). (D) Real-time RT-PCR analysis of the transcript abundance of LINE-1 (L1) mRNAs (ORF1 and ORF2) in normal MCF10A cells and various types of breast cancer cells after being stably transfected with shRNA constructs that encode the endo453 and endo392 sequences. Negative controls containing control shRNA or no shRNA were performed in parallel. The relative LINE-1 mRNA levels were determined after normalizing the data to the control HPRT1 gene. Genomic DNA isolated from HMEC tissue was used as an additional control. Lane 1, parental cells; lane 2, control shRNAs; lane 3, shRNAs encoding endo453; and lane 4, shRNAs encoding endo392. Error bars indicate s.d.
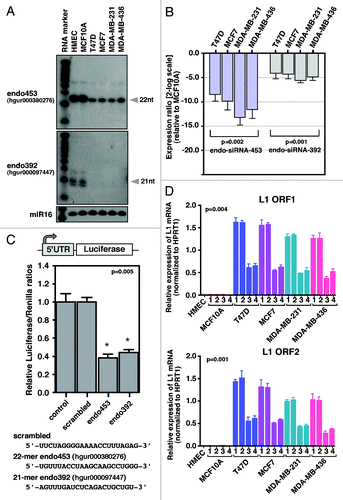
Having identified LINE-1-specific endo-siRNAs, we set out to confirm their inhibitory effect on human LINE-1 expression. To this end, we transfected HEK293T cells with two chemically synthesized endo-siRNAs (21 nt of endo392 and 22 nt of endo453) together with the luciferase reporter gene driven by the LINE-1Hs 5′-UTR promoter and performed a dual luciferase reporter assay (). In this assay, a scrambled endo-siRNA sequence was used as a negative control. The results showed that the transcript levels produced from the LINE-1 promoter were decreased at least 2-fold compared with the control siRNA-transfected cells and parental cells (p = 0.005), suggesting that the identified endo-siRNA species are able to silence LINE-1 expression in human cells.
As the expression of endo-siRNAs is almost undetectable in breast cancer cells, these cells provide an excellent model for investigating the biological functions of endo-siRNA expression. Moreover, breast cancer cells express high levels of LINE-1-encoded transcripts. To further demonstrate that the identified endo-siRNAs were LINE-1 specific, we transfected normal MCF10A and breast cancer cells (T47D, MCF-7, MDA-MB-231 and MDA-MB-436) with two individual shRNA constructs (mimicking endo453 and endo392 sequences) whose expression is driven by constitutively expressing the U6 promoter (Fig. S1). The scrambled endo-siRNA sequence was used as a control, and we performed real-time quantitative RT-PCR as above (). As expected, the overexpression of the endo-siRNAs in pooled cell lines substantially decreased the levels of endogenous LINE-1 mRNAs [both for ORF1 (p = 0.004) and ORF2 (p = 0.001)] by up to 2.5-fold compared with the cells transfected with the scrambled shRNA or non-transfected parental cells and this decrease was strongly correlated with the expression of the endo-siRNAs. Comparison of the LINE-1 retrotransposition frequencies between parental T47D cells and cells overexpressing endo453 showed that the overall percentage of retrotransposition events in the shRNA-expressing cells was 74% lower (average of 963 events vs. 251 events) than that in the parental cells (data not shown). Notably, the overexpression of endo-siRNAs in normal MCF10A breast cells had no effect on LINE-1 expression because MCF10A cells do not express any of the LINE-1-encoded transcripts. These data suggest that the downregulation of endo-siRNAs in cancer cells may contribute to the aberrant expression of LINE-1 elements.
Endo-siRNA increases DNA methylation of the LINE-1 promoter
The exact mechanisms of endo-siRNA action in human LINE-1 silencing are largely unknown. DNA methylation is often associated with the repression of LINE-1 transcription.Citation33 Using a portion of the LINE-1 5′-UTR promoter, several studies have reported an inverse correlation between LINE-1 expression and the methylation status of the LINE-1 promoters.Citation44,Citation45 Recent studies have found that a relatively large group of endo-siRNAs in mice and Drosophila are linked to transposon silencing and DNA methylation.Citation34,Citation37 In addition, the presence of a large CpG island in the human LINE-1 5′-UTR promoter regionCitation46led us to investigate whether the endo-siRNAs could target the DNA methylation of the 5′-UTRs, thereby silencing LINE-1 expression. Surprisingly, a bioinformatics search showed that part of the identified endo453 stem sequence is almost identical to human piRNA-48208 (DQ580096), which controls LINE-1 expression during gametogenesis and cell development, when DNA methylation is not fully active.Citation47
Through an analysis of the sequences of the human LINE-1 promoters using EMBOSS CpGPlot, we identified 31 CpG dinucleotides in the 5′-UTR region forming a CpG island of 534 bp (). The LINE-1 promoter is known to be hypomethylated in several types of human cancer cells activating its expression.Citation32,Citation33 Thus, to examine the methylation status of the LINE-1 5′-UTRs, we initially performed a methylated DNA immunoprecipitation (MeDIP) analysis in parental cells and cells stably expressing the endo-siRNA (endo453 and endo392) sequences. The scrambled endo-siRNA sequence was used as a negative control. The methylated DNAs isolated from the normal MCF10A breast cells and HMECs were used as positive controls. In these assays, two highly conserved sites of the LINE-1 5′-UTRs were examined using two primer sets (I and II) and quantified via a qPCR analysis (). These primer sets were expected to hybridize with most of the LINE-1Hs promoters present in the genome. Compared with the cells transfected with the scrambled siRNA or parental cells, the endo-siRNA-overexpressing cancer cells showed significantly higher levels of DNA methylation, by up to 3-fold for both primer sets (one way ANOVA p < 0.0001 and p < 0.0003 for primer sets I and II, respectively). The extent of DNA methylation varied for each of the primers used because of the global quantification of LINE-1 methylation. Remarkably, the LINE-1 5′-UTR exhibited no methylation changes in normal MCF10A breast cells compared with the same cells treated with the endo-siRNAs. These results clearly suggest that the cells expressing the endo-siRNAs exhibit an overall increase in the levels of LINE-1 DNA methylation.
Figure 6. DNA methylation analysis. (A) Schematic representation of the CpG dinucleotide distribution within the 5′UTR of the LINE-1Hs promoter from +1 to +906 bp (relative to the transcription start site). Vertical lines above indicate the position and numbering of CpG sites; the box represents the CpG island in the LINE-1 promoter (ending at +560 bp). The positions of the primer sets used for methylated DNA immunoprecipitation (MeDIP) and bisulfite sequencing are shown as arrows. (B) MeDIP analysis of LINE-1 5′-UTR methylation patterns. The data are shown as the percentage of methylated DNA relative to the total input for primer sets I and II. Each point represents averages from three experiments. p values were calculated by a one-way analysis of variance. Genomic DNA from HMEC tissue was used as a positive control. The parental cells and cells transfected with control shRNA were analyzed in parallel. Lane 1, parental cells; lane 2, control shRNAs; lane 3, shRNAs encoding endo453; and lane 4, shRNAs encoding endo392. (C) Bisulfite sequence analysis of the LINE-1Hs 5′-UTR region. A region of 560 bp containing 31 CpG sites was sequenced, and the methylation status of individual CpG sites was analyzed within the LINE-1 CpG island. Open and closed circles denote unmethylated and methylated CpG-sites, respectively. Gray represents the unresolved CpG sites. The corresponding percentage of overall CpG methylation is illustrated below, as determined by BDPC analysis.
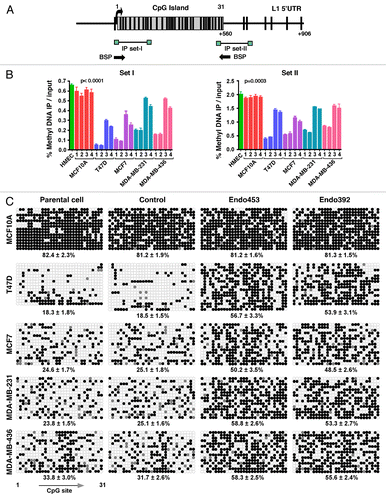
To further investigate the methylation pattern of individual CpG sites in the LINE-1Hs 5′-UTRs, we performed bisulfite conversion analyses on genomic DNA-derived endo-siRNA-treated cells and matched control cells. We amplified a CpG island of the LINE-1 5′-UTR containing 31 CpG sites () and sequenced the resulting amplicons, as described previously.Citation45,Citation48 In practice, this bisulfite primer should amplify all of the methylated LINE-1Hs promoters present in the human genome, thus indicating the global methylation patterns of the LINE-1 promoters. The resulting sequences were analyzed using BiQ-Analyzer software.Citation49 In all cases, we achieved a bisulfite conversion efficiency > 90% (data not shown). Notably, the LINE-1 5′-UTR exhibited significantly higher methylation in cells expressing both of the endo-siRNAs compared with the matched control siRNA-transfected cells or non-transfected parental cells (). An analysis of individual 5′-UTR sequences using BDPC softwareCitation50 showed the greatest variation of LINE-1 CpG methylation between the cells. In all types of cancer cells, the endo-siRNA-treated cells showed significant increases in the level of LINE-1 CpG methylation ranging from 1.6- to 3-fold. These results strongly suggest that the expression of the endo-siRNAs might be related to the methylation status of the CpG islands in the 5′-UTRs. Interestingly, the normal breast MCF10A cells were found to exhibit high CpG methylation (81.5 ± 1.8%) in the 5′-UTR region, irrespective of endo-siRNA expression. Therefore, the absence of LINE-1 expression and de novo LINE-1 retrotransposition events in normal MCF10A cells appear to be due to the high expression of endo-siRNA and the existence of DNA methylation in the promoters. These results demonstrate that endo-siRNAs are required to trigger the overall increases in the DNA methylation of LINE-1 elements in human cells. Our results also imply that depletion of endo-siRNAs is widespread in breast cancer cells and possibly in a range of other breast cancer cells.
Discussion
Breast cancer arises from the mammary epithelium through a multistep sequence of cellular and genetic changes: normal epithelium becomes a breast carcinoma usually by way of hyperplasia, atypical hyperplasia, in situ carcinoma, and invasive carcinoma. Instability of the genome has been suggested to be an important contributor to heritable and genetic changes that drive tumorigenic processes in normal breast cells even before histological abnormalities are detectable.Citation51 The contributions of LINE-1 retrotransposons to pathological processes other than genomic insertions are poorly understood. Earlier studies revealed that LINE-1 is significantly elevated in several types of breast carcinomas,Citation28,Citation29 but the mechanistic pathways that activate LINE-1 expression remain elusive.
Our findings show that non-invasive breast cancer cells express high levels of LINE-1 retrotransposons and their retrotransposition activity compared with moderate-to-invasive cancer cells. At present, it is not clear why LINE-1 is overexpressed in non-invasive cells. One possible explanation for the higher expression is that LINE-1 might become activated in the early stages of the malignant transformation process, even though the onset of tumorigenesis is a multistep process. This early activation of LINE-1 retrotransposon is also in agreement with DNA methylation studies in which the early onset of LINE-1 demethylation has been reported to occur during the progression of many human cancers.Citation52 Our studies also imply that overexpression of LINE-1 may be widespread in breast cancer cells and possibly in a wide range of tumor tissues. Further experiments will determine whether endogenous LINE-1 expression occurs in clinically relevant breast tumors and whether their expression is correlated with the retrotransposition activity.
The expression levels of LINE-1 retrotransposons are extremely low in normal cells, if they are expressed at all. This observation suggests that posttranscriptional control mechanisms are involved in the silencing of LINE-1 expression, presumably via small RNAi-directed transcriptional silencing and recruitment of epigenetic factors to prevent the negative effect of LINE-1 activity within the host genome.Citation37,Citation38 A recent investigation in human cells showed that deletion of a portion of the LINE-1 promoter or knocking down Dicer1 increased LINE-1 expression,Citation16,Citation25 suggesting that siRNAs may be involved in the control of LINE-1 elements, though bona fide siRNAs have yet to be cloned and sequenced. Evidence presented in this study demonstrated, for the first time, the existence of human LINE-1-specific endo-siRNAs that are differentially expressed in normal and breast cancer cells. This study adds to a growing body of evidence indicating that endo-siRNAs repress LINE-1 retrotransposons. Importantly, our finding suggests that endo-siRNAs can silence LINE-1 retrotransposons by increasing DNA methylation, similar to piRNAs linked to retrotransposon methylation in mice.Citation34 This process of gene silencing may be initiated by argonaute protein complexesCitation43 and most likely depends on the recruitment of additional factors to the promoter, such as promoter-specific gene silencing by siRNAs in human cells.Citation38 Studies on Drosophila and S. pombe suggest that some endo-siRNAs, in complex with Ago2 protein, can identify their repeat targets through sequence-specific recognition and thus enabling the targeting of DNA methyltransferase enzymes to the LINE-1 sequences that they modify.Citation34,Citation36 Consistent with this finding, a mutation in Ago2 protein in plants and fission yeasts is correlated with loss of histone H3 Lys9 dimethylation (H3K9me2) and siRNA-directed transcriptional silencing.Citation37 In human cells, targeting promoters with sequence-specific siRNAs leads to transcriptional silencing via histones H3K9me2 and H3K27me3,Citation43 which are characteristic of repressive heterochromatin formation. Although a direct link between endo-siRNAs and repressive histone modifications was not established in this study, the data presented here suggest that endo-siRNAs direct LINE-1 repression through the induction of DNA hypermethylation and possibly through the recruitment of chromatin factors to facilitate these processes. Further experiments are required to determine whether the recruitment of chromatin modifications is required for LINE-1 silencing in human cells.
In mammals, DNA methylation in CpG sites is often closely linked with repressive histone marks that alter chromatin structure, resulting in gene repression. Several studies have found that a loss of LINE-1 DNA methylation is correlated with a loss of genomic stability in cancer cells,Citation32,Citation53 which is a trademark event in tumorigenesis. Interestingly, hypomethylation of the LINE-1 promoter has been reported to be associated with overexpression of LINE-1 transcripts, leading to an accumulation of new insertional mutations upon cancer progression.Citation19 Thus, depletion of endo-siRNAs might lead to DNA hypomethylation in breast cancer cells. Strikingly, our results show that one of the identified endo-siRNAs is endo453 (hgur000380276). Part of this endo453 stem sequence is almost identical to human piRNA-48208 (DQ580096), which controls LINE-1 expression during cellular development.Citation47 At present, it is unknown why endo-siRNAs are downregulated in breast cancer cells and to what extent the expression of endo-siRNAs and their genomic locations are associated with histone modifications. Further studies are required to systematically analyze the epigenetic patterns of endo-siRNA loci and their expression profiles in the human genome. Nevertheless, the data presented in this study demonstrates the following: first, breast cancer cells support the retrotransposition of engineered human LINE-1s in vivo, whereas normal breast cells do not; second, the depletion of LINE-1-specific endo-siRNAs is widespread in breast cancer cells and; third, the overexpression of endo-siRNAs in breast cancer cells increases the levels of CpG methylation in the LINE-1 5′-UTR promoter. Taken together, the results of this study demonstrate that endo-siRNAs are closely associated with LINE-1 expression. This study also adds to a growing body of evidence suggesting that endogenous siRNAs can repress LINE-1 retrotransposons in mammalian cells.
Materials and Methods
Cell cultures
Normal human breast epithelial cells (HMECs) and the MEGM Bullet Kit were obtained from Lonza. The HMEC (Lonza-CC-2551) and immortalized non-tumorigenic MCF10A cells (ATCC-CRL-10371) were cultured using the MEGM Bullet Kit (Clonetics) supplemented with 10 µg/ml of insulin. The MCF7, T47D, MDA-MB-231, and MDA-MB-436 cell lines were obtained from ATCC and cultured in Dulbecco’s modified Eagle’s medium (DMEM) with 2 mM L-glutamine and 10% FCS at 37°C under 5% CO2.
Western blot analysis
Whole-cell lysates were prepared using MPER reagent (Pierce), following the manufacturer’s instructions. Western blot analysis was performed with affinity purified anti-LINE-1-ORF1p antibodiesCitation54 at a 1:5000 dilution (a generous gift from Chris Harris of the University of Medicine and Dentisery of New Jersey), followed by the addition of HRP-conjugated anti-rabbit secondary antibodies (Dako Cytomation). The signals were visualized using the ECL chemiluminescence system (Pierce). To confirm protein normalization, the membranes were stripped and reprobed with mouse α-tubulin antibodies (Sigma).
Quantitative RT-PCR
Total RNA was isolated from cells using the RNeasy Kit (Qiagen) and digested with TurboDNase-I (Ambion). A total of 2 µg of purified RNA was employed for cDNA synthesis with 0.1 µg of random decamer primers using the RETROScript RT Kit (Ambion). The resulting cDNAs were used as templates for qRT-PCR with the following LINE-1-specific and HPRT1 primers: ORF1 forward 5-GGT TAC CCT CAA AGG AAA GCC-3′, ORF1 reverse 5′-GCC TGG TGG TGA CAA AAT CTC-3′; ORF2 forward 5′- AAA TGG TGC TGG GAA AAC TG-3′, ORF2 reverse 5′- GCC ATT GCT TTT GGT GTT TT-3′; HPRT1 forward 5′- CCT GGC GTC GTG ATT AGT GAT-3′, HPRT1 reverse 5′- AGA CGT TCA GTC CTG TCC ATA-3′. The fold changes were determined by comparing the ∆CT value of each product normalized to HPRT1 as an internal control. The data are presented as the average of four independent experiments, with each experiment performed in triplicate. The standard deviations were calculated from the fold changes of the replicates. Unpaired t-tests were used for statistical analysis.
Retrotransposition assay
Exponentially growing normal breast and cancer cells were transfected with human L1RP containing an EGFP retrotransposition cassette (pRP99-L1-EGFP or pRP99-ΔL1-EGFP) using the Amaxa nucleofector kit. Briefly, 1 x 105 cells were suspended in 90 µl of Nucleofection solution, and 10 µl of Nucleofection solution containing 4 µg of LINE-1 vector was added. The mixture was transferred to an electroporation cuvette and electroporated using the A-23 program of the Nucleofector-1 device (Amaxa Biosystems). Approximately 500 µl of pre-warmed medium was added to the cuvette, and the cells were transferred to a 10-cm culture plate. pDsRed-N1 (Clontech) was used as a reporter to examine the transfection efficiency. The transfected cells were enriched by growing them in 200 µg/ml hygromycin. After 9 d of antibiotic selection, untransfected cells were eliminated and maintained in low dose selective medium (75 µg/ml hygromycin). The retrotransposition assay and the determination of retrotransposition frequency were performed at various time points, as described previously.Citation48 The EGFP-positive cells were analyzed by sorting with a FACSVantage DiVa cell sorter (Becton-Dickinson). Over 10,000 individual fluorescent events were acquired using a 530/30 bandpass filter for the EGFP protein signal obtained with fluorescence emission centered at 530 nm. Cells transfected with pRP99-ΔL1-EGFP were used as negative controls to determine background fluorescence throughout the experiment. The percentage of cells that fluoresced below the negative control threshold was considered to be the false positive frequency. Each experimental group was analyzed in triplicate, and two independent transfections per construct were analyzed at each time point.
PCR analysis of retrotransposed cells
Genomic DNA was isolated from cells using a QIAamp DNA kit (Qiagen). PCR was performed to examine the structure of the EGFP reporter cassette integrated with host cell genomic DNA using the Geno-5′ (5′-TAT TGC CGA TCC CCT CAG AAG A-3′) and Geno–3′ (5′-CAA GGA CGA CGG CAA CTA CAA G-3′) primers, as previously described.Citation25The amplified products were visualized on a 1.2% agarose gel. Genomic DNA from retrotransposition-defective (pRP99-ΔL1-EGFP) cells and non-transfected parental cells was used as negative controls.
Small RNA isolation and northern blotting
Approximately 50 µg of total RNA was subjected to enrichment of low MW RNAs by adding 50% PEG-8000 and 5M NaCl to final concentrations of 5% and 0.5 M, respectively. The resulting supernatant was precipitated with ethanol together with 2 µg of glycogen (Promega) and then dissolved in 20 µl of water. Equal amount of the small RNA sample was electrophoresed in a 15% urea-PAGE gel and then transferred to a BrightStar-Plus Nylon membrane (Ambion). DNA oligonucleotide probes detecting LINE-1 sense endo-siRNAs and hsa-miR16 (endo453: 5′-CCC AGG CTT GCT TAG GTA AAC-3′; endo392: 5′-CAG CAG TCT GAG ATC AAA C-3′ and hsa-miR16: 5′-CGC CAA TAT TTA CGT GCT GCT A-3′) were prepared via end labeling with OptiKinase according to the supplier’s instructions (USB, Cleveland, Ohio). Hybridization and washing of the blot were performed as previously described.Citation25 Signals were detected by exposure to X-film for 2–3 d, except for the miR16 probe, which was exposed for only 4 h.
Expression profiling of endo-siRNAs
Small RNA samples were isolated from cells using the mirVanaTM miRNA isolation kit (Ambion) and were reverse-transcribed using custom-made endo-siRNA primers (Applied Biosystems), in accordance with the manufacturer’s protocol. qRT-PCR was performed using TaqMan probes in a 7900HT Thermal cycler (Applied Biosystems) with typical amplification parameters (95°C for 10 min, followed by 40 cycles of 95°C for 15 sec and 60°C for 1 min). The fold changes were determined by comparing the ∆CT value of each endo-siRNA normalized to SnoRA37 (primers obtained from Qiagen). The data generated were the average of three separate experiments, with each experiment performed in triplicate and analyzed using Relative Expression Software (http://REST.gene-quantification.info). For statistical analysis, unpaired t-tests were used.
Luciferase assay
HEK293T cells were cotransfected with modified firefly luciferase (under control of the human LINE-1Hs promoter), a Renilla luciferase reporter plasmid, and chemically synthesized endo-siRNA sequences (obtained from Qiagen). The luciferase assays were performed 48 h after transfection using the Dual-Luciferase Reporter assay system (Promega). The firefly luciferase activity was normalized to the Renilla activity to correct for the transfection efficiency.
Cloning of endo-siRNAs and transfection
shRNAs that mimic 21–22-nt of the endo-siRNAs were designed using the RNAi Codex algorithm (http://katahdin.Cshl.org: 9331/portal/script/main2.pl). A scrambled shRNA was used as a negative control. The templates for each shRNA sequence were produced using a single-step PCR procedure as described previously,Citation55 and cloned directly into the pSM2 vector (Open Biosystem) under the control of the U6 promoter (Fig. S1). To achieve stable transfection, cells (~2 x 105) were seeded into six-well plates and transfected with ~2 µg of shRNA constructs using the Amaxa nucleofector kit (Amaxa Biosystems) and then selected over a one-week period in the presence of 0.6 µg/ml puromycin (Sigma).
Dicer1 and Ago2 shRNA transfection and validation
Dicer1-specific shRNAs (AACCAGGTTGCTCAACAAG, GCTGGCTTATATCAGTAGC) and Ago2-specific shRNAs (CGGCAGGAAGAATCTATAC, GATCGGCAAGAAGAGATTA)Citation41 in the pSiLv-U6 vector were purchased from GeneCopeia (Cat No. HSH006073 and Cat No. HSH007558). As a negative control, an shRNA targeting the luciferase gene in the pSiLv-U6 vector (Cat No. CSHCTR001-LvmU6) was used. Each shRNA was transfected independently into normal MCF10A cells using an Amaxa Nucleofection kit as described above. The depletion of mRNAs was confirmed by qRT-PCR using SYBR Green and 200 nM for forward and reverse primers with the PRISM7700 Sequence Detection System. The primers used were as follows: Dicer1 forward, 5′- TTA ACC TTT TGG TGT TTG ATG AGT GT-3′, Dicer1 reverse, 5′- GCG AGG ACA TGA TGG ACAA TT-3; Ago2 forward, 5′- CTA GAC CCG ACT TTG GGA CCT-3′, Ago2 reverse, 5′- GGG CAC TTC TCT GGT TGA TA-3′. The measured transcript levels were normalized to HPRT1. The samples were amplified in triplicate. For protein analysis, western blotting of whole-cell lysates was performed using affinity-purified anti-Dicer1 and Ago2 (Abcam) antibodies at 1:1000 and 1:500 dilutions, respectively, followed by the addition of HRP-conjugated secondary antibodies. The resultant signals were visualized using the ECL chemiluminescence system (Pierce). To confirm protein normalization, the membranes were stripped and reprobed with α-tubulin antibodies (Sigma).
Methylated DNA immunoprecipitation (MeDIP)
Genomic DNA was extracted from cells using the Qiagen DNA Mini kit (Qiagen). Approximately 2 µg of DNA was sonicated in 200 µl of H2O for ten sonication cycles (30 sec “on” and 30 sec “off”) using a Biorupter sonicator (Diagenode) to produce sheared DNA with lengths of 300–1000 bp. Methylated DNA was precipitated from 1 µg of the sheared DNA using 1 µg of an anti-5-methylcytosine antibody (Epigentek) according to the manufacturer’s instructions. As a background control, precipitation was performed using a normal mouse IgG antibody. The precipitated DNA was quantified by qPCR using the two following sets of the primers: Set-I forward, 5′-ACA GGA ACT GCG GTG GAG-3′, Set-I reverse, 5′-AAC TCC CTG ACC CCT TGC-3′; Set-II forward, 5′-GGC ACA CTG ACA CCT CAC C-3′, Set-II reverse, 5′-TGG TCT TTG ATG ATG GTG AT-3′. The graph represents the average of three replicate qPCR reactions for precipitated DNA and input DNA. The calculated errors in all graphs are the standard deviations from triplicate reactions. For the statistical analysis, a one-way ANOVA was used.
Bisulphite sequencing analysis
After isolating genomic DNA as described above, 4 µg of DNA was treated with the Methyl-Easy DNA bisulfite modification kit (Human Genetic Signatures) according the manufacturer’s instructions. The primers used for bisulfite sequencing of the LINE-1 promoter were designed using Methyl Primer Express software (Applied Biosystems). A forward primer (5′-TTA TAA ATT ATG TTT TTG TGA ATG GAT AGT-3′) and a reverse primer (5′-TAA TTT TAT CTA CTT TTA ATC TTT AAT AAT-3′) that bind conserved sequences of LINE-1 promoters were used for this assay. Approximately 60 ng of bisulfite-modified DNA was subjected to PCR using the following conditions: initial denaturation at 94°C for 3 min, followed by 35 cycles of 94°C for 1 min, 52°C for 2 min and 72°C for 2 min and a final step by 72°C for 10 min. The obtained amplicons were analyzed on 1.5% agarose gels and isolated with the MinElute Gel extraction kit (Qiagen). The purified PCR products were cloned into the pGEM-T easy vector (Promega) and sequenced as described previously.Citation45 The sequence of each amplicon was analyzed with using BiQ Analyzer software (http://biq-analyzer.bioinf.mpi-inf.mpg.de) to assess sequence quality and to visualize the DNA methylation data. The average methylation levels and the percentage of each CpG site were analyzed by the using BDPC web server (http://biochem.jacobs-university.de/BDPC).
| Abbreviations: | ||
| LINE | = | long interspersed nuclear element |
| SINE | = | short interspersed nuclear element |
| endo-siRNA | = | endogenous small interfering RNA |
Additional material
Download Zip (744.1 KB)Acknowledgments
This work was supported by a grant from the Australian National Health and Medical Research Council (418021) and the ACT Health and Medical Research Support Program.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Brouha B, Schustak J, Badge RM, Lutz-Prigge S, Farley AH, Moran JV, et al. Hot L1s account for the bulk of retrotransposition in the human population. Proc Natl Acad Sci U S A 2003; 100:5280 - 5; http://dx.doi.org/10.1073/pnas.0831042100; PMID: 12682288
- Beck CR, Collier P, Macfarlane C, Malig M, Kidd JM, Eichler EE, et al. LINE-1 retrotransposition activity in human genomes. Cell 2010; 141:1159 - 70; http://dx.doi.org/10.1016/j.cell.2010.05.021; PMID: 20602998
- Scott AF, Schmeckpeper BJ, Abdelrazik M, Comey CT, O’Hara B, Rossiter JP, et al. Origin of the human L1 elements: proposed progenitor genes deduced from a consensus DNA sequence. Genomics 1987; 1:113 - 25; http://dx.doi.org/10.1016/0888-7543(87)90003-6; PMID: 3692483
- Martin SL, Bushman FD. Nucleic acid chaperone activity of the ORF1 protein from the mouse LINE-1 retrotransposon. Mol Cell Biol 2001; 21:467 - 75; http://dx.doi.org/10.1128/MCB.21.2.467-475.2001; PMID: 11134335
- Feng Q, Moran JV, Kazazian HH Jr., Boeke JD. Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell 1996; 87:905 - 16; http://dx.doi.org/10.1016/S0092-8674(00)81997-2; PMID: 8945517
- Mathias SL, Scott AF, Kazazian HH Jr., Boeke JD, Gabriel A. Reverse transcriptase encoded by a human transposable element. Science 1991; 254:1808 - 10; http://dx.doi.org/10.1126/science.1722352; PMID: 1722352
- Moran JV, Holmes SE, Naas TP, DeBerardinis RJ, Boeke JD, Kazazian HH Jr.. High frequency retrotransposition in cultured mammalian cells. Cell 1996; 87:917 - 27; http://dx.doi.org/10.1016/S0092-8674(00)81998-4; PMID: 8945518
- Goodier JL, Kazazian HH Jr.. Retrotransposons revisited: the restraint and rehabilitation of parasites. Cell 2008; 135:23 - 35; http://dx.doi.org/10.1016/j.cell.2008.09.022; PMID: 18854152
- Cordaux R, Batzer MA. The impact of retrotransposons on human genome evolution. Nat Rev Genet 2009; 10:691 - 703; http://dx.doi.org/10.1038/nrg2640; PMID: 19763152
- Esnault C, Maestre J, Heidmann T. Human LINE retrotransposons generate processed pseudogenes. Nat Genet 2000; 24:363 - 7; http://dx.doi.org/10.1038/74184; PMID: 10742098
- Terai G, Yoshizawa A, Okida H, Asai K, Mituyama T. Discovery of short pseudogenes derived from messenger RNAs. Nucleic Acids Res 2010; 38:1163 - 71; http://dx.doi.org/10.1093/nar/gkp1098; PMID: 19965772
- Ostertag EM, Kazazian HH Jr.. Biology of mammalian L1 retrotransposons. Annu Rev Genet 2001; 35:501 - 38; http://dx.doi.org/10.1146/annurev.genet.35.102401.091032; PMID: 11700292
- Belancio VP, Roy-Engel AM, Pochampally RR, Deininger P. Somatic expression of LINE-1 elements in human tissues. Nucleic Acids Res 2010; 38:3909 - 22; http://dx.doi.org/10.1093/nar/gkq132; PMID: 20215437
- Kano H, Godoy I, Courtney C, Vetter MR, Gerton GL, Ostertag EM, et al. L1 retrotransposition occurs mainly in embryogenesis and creates somatic mosaicism. Genes Dev 2009; 23:1303 - 12; http://dx.doi.org/10.1101/gad.1803909; PMID: 19487571
- Iskow RC, McCabe MT, Mills RE, Torene S, Pittard WS, Neuwald AF, et al. Natural mutagenesis of human genomes by endogenous retrotransposons. Cell 2010; 141:1253 - 61; http://dx.doi.org/10.1016/j.cell.2010.05.020; PMID: 20603005
- Yang N, Kazazian HH Jr.. L1 retrotransposition is suppressed by endogenously encoded small interfering RNAs in human cultured cells. Nat Struct Mol Biol 2006; 13:763 - 71; http://dx.doi.org/10.1038/nsmb1141; PMID: 16936727
- Garcia-Perez JL, Morell M, Scheys JO, Kulpa DA, Morell S, Carter CC, et al. Epigenetic silencing of engineered L1 retrotransposition events in human embryonic carcinoma cells. Nature 2010; 466:769 - 73; http://dx.doi.org/10.1038/nature09209; PMID: 20686575
- Harris CR, Normart R, Yang Q, Stevenson E, Haffty BG, Ganesan S, et al. Association of nuclear localization of a long interspersed nuclear element-1 protein in breast tumors with poor prognostic outcomes. Genes Cancer 2010; 1:115 - 24; http://dx.doi.org/10.1177/1947601909360812; PMID: 20948976
- Roman-Gomez J, Jimenez-Velasco A, Agirre X, Cervantes F, Sanchez J, Garate L, et al. Promoter hypomethylation of the LINE-1 retrotransposable elements activates sense/antisense transcription and marks the progression of chronic myeloid leukemia. Oncogene 2005; 24:7213 - 23; http://dx.doi.org/10.1038/sj.onc.1208866; PMID: 16170379
- Aporntewan C, Phokaew C, Piriyapongsa J, Ngamphiw C, Ittiwut C, Tongsima S, et al. Hypomethylation of intragenic LINE-1 represses transcription in cancer cells through AGO2. PLoS One 2011; 6:e17934; http://dx.doi.org/10.1371/journal.pone.0017934; PMID: 21423624
- Wolff EM, Byun HM, Han HF, Sharma S, Nichols PW, Siegmund KD, et al. Hypomethylation of a LINE-1 promoter activates an alternate transcript of the MET oncogene in bladders with cancer. PLoS Genet 2010; 6:e1000917; http://dx.doi.org/10.1371/journal.pgen.1000917; PMID: 20421991
- Okamura K, Lai EC. Endogenous small interfering RNAs in animals. Nat Rev Mol Cell Biol 2008; 9:673 - 8; http://dx.doi.org/10.1038/nrm2479; PMID: 18719707
- Kawamura Y, Saito K, Kin T, Ono Y, Asai K, Sunohara T, et al. Drosophila endogenous small RNAs bind to Argonaute 2 in somatic cells. Nature 2008; 453:793 - 7; http://dx.doi.org/10.1038/nature06938; PMID: 18463636
- Watanabe T, Totoki Y, Toyoda A, Kaneda M, Kuramochi-Miyagawa S, Obata Y, et al. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature 2008; 453:539 - 43; http://dx.doi.org/10.1038/nature06908; PMID: 18404146
- Lee SH, Eldi P, Cho SY, Rangasamy D. Control of chicken CR1 retrotransposons is independent of Dicer-mediated RNA interference pathway. BMC Biol 2009; 7:53; http://dx.doi.org/10.1186/1741-7007-7-53; PMID: 19691826
- Nilsen TW. Endo-siRNAs: yet another layer of complexity in RNA silencing. Nat Struct Mol Biol 2008; 15:546 - 8; http://dx.doi.org/10.1038/nsmb0608-546; PMID: 18523465
- Moazed D. Small RNAs in transcriptional gene silencing and genome defence. Nature 2009; 457:413 - 20; http://dx.doi.org/10.1038/nature07756; PMID: 19158787
- Bratthauer GL, Cardiff RD, Fanning TG. Expression of LINE-1 retrotransposons in human breast cancer. Cancer 1994; 73:2333 - 6; http://dx.doi.org/10.1002/1097-0142(19940501)73:9<2333::AID-CNCR2820730915>3.0.CO;2-4; PMID: 8168038
- Asch HL, Eliacin E, Fanning TG, Connolly JL, Bratthauer G, Asch BB. Comparative expression of the LINE-1 p40 protein in human breast carcinomas and normal breast tissues. Oncol Res 1996; 8:239 - 47; PMID: 8895199
- Dmitriev SE, Andreev DE, Terenin IM, Olovnikov IA, Prassolov VS, Merrick WC, et al. Efficient translation initiation directed by the 900-nucleotide-long and GC-rich 5′ untranslated region of the human retrotransposon LINE-1 mRNA is strictly cap dependent rather than internal ribosome entry site mediated. Mol Cell Biol 2007; 27:4685 - 97; http://dx.doi.org/10.1128/MCB.02138-06; PMID: 17470553
- Leibold DM, Swergold GD, Singer MF, Thayer RE, Dombroski BA, Fanning TG. Translation of LINE-1 DNA elements in vitro and in human cells. Proc Natl Acad Sci U S A 1990; 87:6990 - 4; http://dx.doi.org/10.1073/pnas.87.18.6990; PMID: 1698287
- Hoffmann MJ, Schulz WA. Causes and consequences of DNA hypomethylation in human cancer. Biochemistry and cell biology 2005; 83:296 - 321
- Perrin D, Ballestar E, Fraga MF, Frappart L, Esteller M, Guerin JF, et al. Specific hypermethylation of LINE-1 elements during abnormal overgrowth and differentiation of human placenta. Oncogene 2007; 26:2518 - 24; http://dx.doi.org/10.1038/sj.onc.1210039; PMID: 17043645
- Aravin AA, Sachidanandam R, Bourc’his D, Schaefer C, Pezic D, Toth KF, et al. A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol Cell 2008; 31:785 - 99; http://dx.doi.org/10.1016/j.molcel.2008.09.003; PMID: 18922463
- Watanabe T, Takeda A, Tsukiyama T, Mise K, Okuno T, Sasaki H, et al. Identification and characterization of two novel classes of small RNAs in the mouse germline: retrotransposon-derived siRNAs in oocytes and germline small RNAs in testes. Genes Dev 2006; 20:1732 - 43; http://dx.doi.org/10.1101/gad.1425706; PMID: 16766679
- Ghildiyal M, Seitz H, Horwich MD, Li C, Du T, Lee S, et al. Endogenous siRNAs derived from transposons and mRNAs in Drosophila somatic cells. Science 2008; 320:1077 - 81; http://dx.doi.org/10.1126/science.1157396; PMID: 18403677
- Malone CD, Hannon GJ. Small RNAs as guardians of the genome. Cell 2009; 136:656 - 68; http://dx.doi.org/10.1016/j.cell.2009.01.045; PMID: 19239887
- Girard A, Hannon GJ. Conserved themes in small-RNA-mediated transposon control. Trends Cell Biol 2008; 18:136 - 48; http://dx.doi.org/10.1016/j.tcb.2008.01.004; PMID: 18282709
- Speek M. Antisense promoter of human L1 retrotransposon drives transcription of adjacent cellular genes. Mol Cell Biol 2001; 21:1973 - 85; http://dx.doi.org/10.1128/MCB.21.6.1973-1985.2001; PMID: 11238933
- Czech B, Malone CD, Zhou R, Stark A, Schlingeheyde C, Dus M, et al. An endogenous small interfering RNA pathway in Drosophila. Nature 2008; 453:798 - 802; http://dx.doi.org/10.1038/nature07007; PMID: 18463631
- Zhou Y, Chen L, Barlogie B, Stephens O, Wu X, Williams DR, et al. High-risk myeloma is associated with global elevation of miRNAs and overexpression of EIF2C2/AGO2. Proc Natl Acad Sci U S A 2010; 107:7904 - 9; http://dx.doi.org/10.1073/pnas.0908441107; PMID: 20385818
- Yang JH, Shao P, Zhou H, Chen YQ, Qu LH. deepBase: a database for deeply annotating and mining deep sequencing data. Nucleic Acids Res 2010; 38:Database issue D123 - 30; http://dx.doi.org/10.1093/nar/gkp943; PMID: 19966272
- Kim DH, Villeneuve LM, Morris KV, Rossi JJ. Argonaute-1 directs siRNA-mediated transcriptional gene silencing in human cells. Nat Struct Mol Biol 2006; 13:793 - 7; http://dx.doi.org/10.1038/nsmb1142; PMID: 16936726
- Bourc’his D, Bestor TH. Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3L. Nature 2004; 431:96 - 9; http://dx.doi.org/10.1038/nature02886; PMID: 15318244
- Coufal NG, Garcia-Perez JL, Peng GE, Yeo GW, Mu Y, Lovci MT, et al. L1 retrotransposition in human neural progenitor cells. Nature 2009; 460:1127 - 31; http://dx.doi.org/10.1038/nature08248; PMID: 19657334
- Hata K, Sakaki Y. Identification of critical CpG sites for repression of L1 transcription by DNA methylation. Gene 1997; 189:227 - 34; http://dx.doi.org/10.1016/S0378-1119(96)00856-6; PMID: 9168132
- Girard A, Sachidanandam R, Hannon GJ, Carmell MA. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature 2006; 442:199 - 202; PMID: 16751776
- Rangasamy D. An S/MAR-based L1 retrotransposition cassette mediates sustained levels of insertional mutagenesis without suffering from epigenetic silencing of DNA methylation. Epigenetics 2010; 5:601 - 11; http://dx.doi.org/10.4161/epi.5.7.12647; PMID: 20595816
- Bock C, Reither S, Mikeska T, Paulsen M, Walter J, Lengauer T. BiQ Analyzer: visualization and quality control for DNA methylation data from bisulfite sequencing. Bioinformatics 2005; 21:4067 - 8; http://dx.doi.org/10.1093/bioinformatics/bti652; PMID: 16141249
- Rohde C, Zhang Y, Jurkowski TP, Stamerjohanns H, Reinhardt R, Jeltsch A. Bisulfite sequencing Data Presentation and Compilation (BDPC) web server--a useful tool for DNA methylation analysis. Nucleic Acids Res 2008; 36:e34; http://dx.doi.org/10.1093/nar/gkn083; PMID: 18296484
- Ellsworth DL, Ellsworth RE, Liebman MN, Hooke JA, Shriver CD. Genomic instability in histologically normal breast tissues: implications for carcinogenesis. Lancet Oncol 2004; 5:753 - 8; http://dx.doi.org/10.1016/S1470-2045(04)01653-5; PMID: 15581548
- Sunami E, de Maat M, Vu A, Turner RR, Hoon DS. LINE-1 hypomethylation during primary colon cancer progression. PLoS One 2011; 6:e18884; http://dx.doi.org/10.1371/journal.pone.0018884; PMID: 21533144
- Phokaew C, Kowudtitham S, Subbalekha K, Shuangshoti S, Mutirangura A. LINE-1 methylation patterns of different loci in normal and cancerous cells. Nucleic Acids Res 2008; 36:5704 - 12; http://dx.doi.org/10.1093/nar/gkn571; PMID: 18776216
- Harris CR, Normart R, Yang Q, Stevenson E, Haffty BG, Ganesan S, et al. Association of nuclear localization of a long interspersed nuclear element-1 protein in breast tumors with poor prognostic outcomes. Genes Cancer 2010; 1:115 - 24; http://dx.doi.org/10.1177/1947601909360812; PMID: 20948976
- Rangasamy D, Tremethick DJ, Greaves IK. Gene knockdown by ecdysone-based inducible RNAi in stable mammalian cell lines. Nat Protoc 2008; 3:79 - 88; http://dx.doi.org/10.1038/nprot.2007.456; PMID: 18193024