Abstract
Methyl-CpG binding protein 2 (MeCP2) binds methylated cytosines at CpG sites on DNA and it is thought to function as a critical epigenetic regulator. Mutations in the MeCP2 gene have been associated to Rett syndrome, a human neurodevelopmental disorder. Here we show that MeCP2 is acetylated by p300 and that SIRT1 mediates its deacetylation. SIRT1, the mammalian homologue of Sir2 in yeast, is a nicotinamide-adenine dinucleotide (NAD+)-dependent histone deacetylase that belongs to the family of HDAC class III sirtuins. Importantly, SIRT1 has been shown to play a critical role in synaptic plasticity and memory formation. This study reveals a functional interplay between two critical epigenetic regulators, MeCP2 and SIRT1, which controls MeCP2 binding activity to the brain-derived neurotrophic factor (BDNF) promoter in a specific region of the brain.
Keywords: :
Introduction
Methyl-CpG binding protein 2 (MeCP2) is a very abundant protein that was identified for its capacity to bind methylated cytosines at CpG sites.Citation1,Citation2 MeCP2 is highly conserved within mammals and it contains a N-terminal methyl-CpG-binding domain (MBD) and a C-terminal transcriptional repressor domain (TRD).Citation3 A large body of evidence has established the concept that DNA methylation is associated with transcriptional silencing. The family of MBD binding proteins has been intimately linked to this process.Citation4 Moreover, MeCP2 is capable of further inhibiting transcription through the recruitment of Sin3A and histone deacetylases.Citation3 MeCP2 expression is present in many different tissues, but it appears to be most abundant in the brain.Citation5 The MeCP2 gene, located on the X-chromosome, is transcribed into two different splicing isoforms, MeCP2e1 and MeCP2e2, that differ only in a few amino acids of their N-terminal regions.Citation6 While the e2 isoform was the first one to be identified, the e1 isoform is the most expressed in the brain in both human and mice.Citation7,Citation8 Mutations in the MeCP2 gene have been associated with Rett syndrome,Citation9,Citation10 a neurodevelopmental disorder with affected individuals displaying autistic features, mental retardation and motor and respiratory abnormalities.Citation11 MeCP2 is capable of controling the expression of several genes that play a crucial role during the process of synapse formation, such as brain-derived neurotrophic factor (BDNF), inhibitors of differentiation (IDs), early growth gene response 2 (EGR2) and JunB.Citation12-Citation14 The molecular and physiological pathways controlling MeCP2 function have remained elusive.
SIRT1, the mammalian homologue of Sir2 in yeast, is a nicotinamide-adenine dinucleotide (NAD+)-dependent histone deacetylase.Citation15 Its function is tightly coupled to cellular metabolism and its dysfunction has been linked to inflammation, obesity and cancer.Citation15 Accumulating evidence underscores the importance of SIRT1-mediated epigenetic control in neuronal plasticity.Citation16,Citation17 It has been described that SIRT1 can regulate neuronal differentiationCitation18,Citation19 and also prevents neurodegeneration in mouse models of Alzheimer’s disease.Citation20-Citation22 Moreover, Tau acetylation can be reverted by SIRT1.Citation23 Interestingly, SIRT1 brain-specific knockout mice show impaired cognitive abilities.Citation24 This enzyme can also regulate synaptic plasticity and memory formation through a microRNA-mediated mechanism.Citation25
In this study, we show that MeCP2 is acetylated at a specific lysine residue and that SIRT1 mediates its reversible deacetylation. This post-translational modification appears to modulate MeCP2 binding at the BDNF promoter in the hippocampus. Thus, we reveal the convergence in the control of SIRT1 and MeCP2, two critical epigenetic regulators.
Results
MeCP2 protein is acetylated by p300
Through a high-resolution mass spectrometry screen, it was found that the MeCP2 protein is acetylated at a single site corresponding to lysine 461 of human MeCP2.Citation26 In order to investigate if acetylation could operate as a post-translational modification that regulates MeCP2 function, we first determined if this lysine was conserved between different species. As shown in Figure S1A, the putative acetylated lysine is conserved across several species, and it corresponds to lysine 464 of the mouse e1 isoform, which is the most expressed MeCP2 isoform in the brain. We found that the MeCP2e1 isoform is readily acetylated in vitro (). To determine whether MeCP2 is acetylated in vivo, we generated a polyclonal antibody that specifically recognizes the acetylated form of MeCP2e1 when modified at lysine 464. We ectopically expressed MeCP2e1 or a mutant with the single amino acid K464R conversion in HEK-293 cells. The K464R mutation mimics a deacetylated lysine residue and is not recognizable by the anti-acetyl MeCP2 antibody (Fig. S1B). Analyzing the primary sequence of the MeCP2e1 protein, we detected several p300 binding motifs.Citation27 We then sought to investigate if this acetyltransferase could mediate MeCP2 acetylation in vitro. We co-transfected p300 or CBP together with MeCP2e1 or the MeCP2e1-K464 point mutant in HEK-293 cells. Co-expression of p300 leads to an efficient increase in the levels of MeCP2 acetylation at lysine 464 (). On the contrary, CBP is not able to efficiently induce acetylation of MeCP2. It should be noted that, even when using equivalent expression vectors, CBP is always expressed at lower levels when compared to p300 (). To definitely demonstrate that MeCP2 is acetylated, we purified mouse recombinant MeCP2 with glutathione S-transferase (GST) or a control GST-empty, and MeCP2 acetylation was detected only in the presence of purified recombinant p300 (). In addition, in vitro incorporation assays using 14C-acetyl-Coenzyme A demonstrated that the lysine 464 is acetylated upon addition of p300 ().
Figure 1. MeCP2 is acetylated by p300 at Lys-464. (A) The mouse MeCP2 isoform 1 (MeCP2e1-FLAG) expression vector was transfected in HEK-293 cells, and immunoprecipitated samples were probed with anti-pan-acetyl-lysine antibody. (B) HA-tagged p300, but not HA-tagged CBP, increases acetylation of MeCP2 at lysine 464 in HEK-293 cells. FLAG-MeCP2e1 WT or FLAG-MeCP2e1 K464 point mutant were co-expressed together with HA-tagged p300 or HA-tagged CBP in HEK-293 cells. (C) Level of acetylated Lys-464/ MeCP2 was set as 1; p300 is capable to significantly increase the amount of acetylated MeCP2. Blots are representative of three independent experiments (n=3). Student's t test. Values represent means ± SD. *p<0.02 **p<0.01 (D) Anti-acetyl MeCP2-K464 (AcMeCP2) antibody recognizes recombinant GST-MeCP2e1 acetylated by p300, not GST-MeCP2e1 alone or GST-empty (GST-EV). (E) In vitro acetylation assay. Signal for acetylated MeCP2 is detected in the presence of p300. (F-G) MeCP2 interacts with wild type SIRT1, but not with the catalytically inactive mutant H363Y (HEK-293). (H) SIRT1 wild type (WT) or the catalytically inactive mutant (H363Y) were co-expressed in HEK-293 cells together with MeCP2e1. After MeCP2 immunoprecipitation the level of acetylated Lys-464 was monitored in the presence of SIRT1 WT or H363Y mutant. (I) Level of acetylated Lys-464/ MeCP2 was set as 1. Co-expression of SIRT1 WT, but not the catalytically inactive one (H363Y), decreases K464 acetylation. Blots are representative of three independent experiments (n=3). Student's t test *p<0.02 **p<0.01. t test n=3. Values represent means ± SD. *p<0.02 **p<0.01
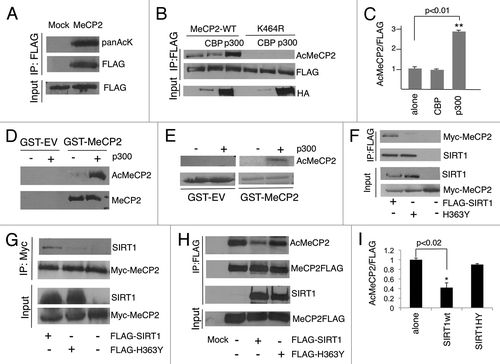
SIRT1 mediates MeCP2 deacetylation
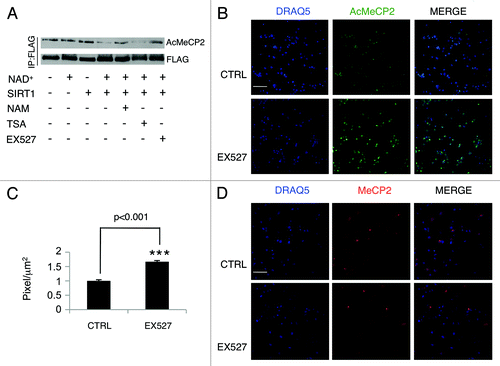
The evidence that MeCP2 undergoes acetylation implies that a deacetylase may reverse this post-translational modification. SIRT1 involvement in cognition, memory formation and neurodegenerative disorders has recently been described.Citation17,Citation19,Citation20,Citation24 In addition, SIRT1 has been shown to readily deacetylate and interact with various non-histone substrates.Citation15,Citation17 Thus, we sought to investigate whether SIRT1 interacts with MeCP2. When co-expressed in HEK-293 cells, MeCP2 readily co-immunoprecipitates with SIRT1 (). Strikingly, MeCP2 does not interact with the SIRT1 mutant H363Y, in which a single amino acid is converted in order to impair its deacetylase enzymatic activity.Citation28 This suggests that SIRT1 enzymatic function is required to induce formation of the SIRT1-MeCP2 complex. Moreover, the MeCP2 point mutant K464R exclusively interacts with SIRT1 (Fig. S2A). Importantly, other members of the sirtuin family failed to interact with MeCP2e1, underscoring the specificity of the SIRT1-MeCP2 interaction (Fig. S2B). In order to map the region of interaction between SIRT1 and MECP2e1, we used different MeCP2e1 truncation mutants (Fig. S3A). The C-terminal portion of the protein is required and sufficient for interaction with SIRT1 (Fig. S3B). To determine whether SIRT1 induces MeCP2 deacetylation, we co-expressed MePC2e1 with SIRT1, or its enzymatically inactive H363Y mutant. SIRT1 readily induces deacetylation at lysine 464 (). Subsequently, we sought to investigate whether SIRT1 could deacetylate MeCP2e1 in an in vitro deacetylation assay. As shown in , SIRT1 induces K464 deacetylation in the presence of the co-factor NAD+. When nicotinamide (NAM) is added, the acetylation levels of MeCP2 increase, as expected by the inhibitory effect that NAM has on SIRT1. The same effect was observed when EX527, a SIRT1 specific inhibitor, was added. This result confirms the capacity of SIRT1 to deacetylate MeCP2 at the targeted lysine in vitro.
Figure 2. SIRT1 controls MeCP2 acetylation in vitro and in primary neurons. (A) SIRT1 deacetylates MeCP2 in vitro. Immunoprecipitated MeCP2 was incubated with purified recombinant SIRT1 in deacetylation buffer that contained NAD+, TSA, EX527, or nicotinamide. MeCP2 acetylation at lysine 464 (AcMeCP2) was monitored by Western analysis. The experiments were repeated three times (B) Immunofluorescence of cortical neurons (DIV5) after EX527 treatment (6 hours). Bar=65 mm (C) Intensity of fluorescence shown in panel B is evaluated as pixels/µm2 (Materials and Methods) Student's t test (n=3) Values represent means ± SD. *p<0.05, **p<0.01, ***p<0.001. (D) Immunofluorescence of cortical neurons (DIV5) after EX527 treatment (6 hours). Cells were stained with anti-acetyl MeCP2 or anti-MeCP2 antibody. Bar=65 mm
MeCP2 acetylation and its effect on BDNF expression
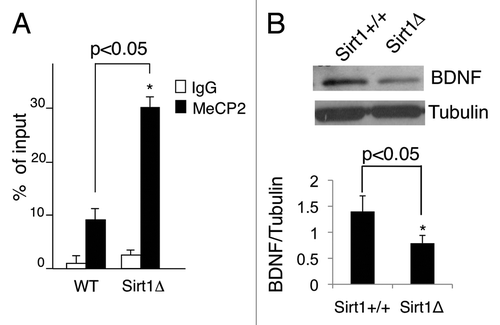
To determine whether MeCP2 acetylation could be detected in vivo, primary cortical neurons were prepared from newborn mice. After 5 days in culture (DIV5), immunofluorescence experiments using the anti-acetyl MeCP2 antibody were performed. As shown in , a clear nuclear signal was detected, demonstrating that acetylation of MeCP2 at K464 occurs in vivo. Importantly, treatment with the SIRT1-specific inhibitor EX527 increased acetylation significantly (~50%) (). The total amount of MeCP2 protein remains unchanged after the treatment with EX527 (). These results confirmed that acetylation of MeCP2 is controlled by SIRT1. Consequently, we sought to investigate whether MeCP2 acetylation could modify its recruitment to its primary target, the BDNF promoter. Chromatin immunoprecipitation (ChIP) analyses performed on fresh hippocampus tissue showed that MeCP2 recruitment on the BDNF exon 4 promoter was significantly higher in SIRT1Δex4 mice as compared to the wild type littermates (). This difference in chromatin recruitment was associated to a decrease in both protein () and mRNA levels of BDNF in SIRT1Δex4 mice, in accordance to previous reports.Citation25
Figure 3. (A) MeCP2 binding to the BDNF exon IV promoter (200 bp upstream from the transcriptional starting site) is increased in SIRT1Δex4/Nestin-Cre hippocampi as revealed by ChIP analysis (n=3 animals per each genotype). Student's t-test. Values represent means ± SD. *p<0.05, **p<0.01, ***p<0.001. (B) Reduced BDNF protein level in SIRT1Δex4 hippocampi. Level of BDNF mRNA in SIRT1 wild type animals was set as control. Student's t-test (n=3) Values represent means ± SD. *p<0.05, **p<0.01, ***p<0.001.
Conclusion
Methyl-CpG binding protein 2 (MeCP2) is capable of binding methylated cytosines at CpG sites, and mutations in its gene have been associated with Rett Syndrome. It has been previously described that post-translational modifications can regulate MeCP2 protein function. Phosphorylation at serine 80 or serine 421 regulate, in opposite ways, the binding or release of MeCP2 from BDNF gene promoter.Citation29,Citation30 In this study, we demonstrated that MeCP2 undergoes lysine acetylation, an event mediated by the acetyltransferase p300 in vitro. More interestingly, SIRT1, a NAD+-dependent histone deacetylase, appears to mediate the opposite reversible reaction. Importantly, accumulating evidence underscores the importance of this histone deacetylase in regulating neuronal differentiation.Citation31,Citation32
We speculate that SIRT1-dependent deacetylation of MeCP2 could allow its release from the methylated CpG sites within the BDNF exon 4 promoter leading to increased BDNF transcription. In keeping with this hypothesis, the absence of a functional SIRT1 in SIRT1Δex4 mice prevents the release of the acetylated MeCP2 from the BDNF promoter, resulting in decreased expression.
Our findings indicate that control of BDNF expression may implicate the interplay of two epigenetic pathways, where SIRT1-mediated deacetylation appears to influence MeCP2 binding to methylated DNA. Due to the strong connection between SIRT1 and metabolism, this scenario also contemplates the intriguing possibility that MeCP2 function may be modulated by changes in the metabolic state of specific neurons.
Materials and Methods
Mice
The SIRT1Dex4/Nestin-Cre mice and the SIRT1 +/+Nestin-Cre mice were kindly provided by Dr. L.H. Tsai.
Primary neuronal cultures
Primary cortical neurons were prepared from wild type pups (1-2 postnatal days). Purified cells were plated using Neurobasal A supplemented with B27 (Invitrogen) on poly-D-lysine coated plates. All the experiments were performed at 5 days in vitro culture (DIV).
Chromatin immunoprecipitation (ChIP)
Hippocampi were dissected from SIRT1Dex4/Nestin-Cre brain and disuccinimidyl glutarate (DSG) was added to a final concentration of 2 mM for crosslinking. After 45 minutes at room temperature, formaldehyde was added to a final concentration of 1% (v/v) and cells incubated for 15 min. After dual crosslinking, glycine was added to a final concentration of 0.1M and incubated for 10 min to quench formaldehyde. Samples were homogenized, resuspended in lysis buffer and sonicated to shear DNA. The whole-cell extract was incubated over-night at 4°C with the appropriate antibody. Protein G/salmon sperm was added and after two hours beads were washed one time in lysis buffer, one time in low salt buffer (150 mM NaCl), one time in high salt buffer (500 mM NaCl), one time in LiCl, and two times in TE buffer. Bound complexes were eluted from the beads with elution buffer (10%SDS, 0.1M NaHCO3) followed by heating at 65°C overnight (reverse cross-linking). Immunoprecipitated complexes were treated with RNase A, proteinase K, and phenol:chloroform:isoamyl alcohol extraction. Purified DNA samples were normalized and subjected to real-time PCR. Sequence of BDNF primersCitation12: BDNF exon IV -200 FW: 5’-GGC TTC TGT GTG CGT GAA TTT GC-3’; BDNF exon IV 0 REW: 5’- AAA GTG GGT GGG AGT CCA CGA G-3’
Immunofluorescence and quantification
Cells were fixed using 4% paraformaldehyde in PBS. Blocking was performed using 3% BSA diluted in PBS for 30 minutes at room temperature. Primary antibody used: anti-acetyl MeCP2 (ABE28 Millipore 1: 400 dilution) was incubated overnight. Secondary antibody was conjugated with Alexa-488 goat anti-rabbit (Invitrogen). Nuclei were stained with DRAQ-5 [-(dimethylamino)ethylamino-4,8-dihydroxyanthracene-9,10-dione] (Biostatus Limited). Immunolabeled sections were examined with a Leica confocal microscope SP5 (DMRE, Leica). Controls were always performed by omitting primary antibodies. Intensity of fluorescence is evaluated as pixels/µm2. The Leica SP5 software LAS AF was used for quantification.
In vitro deacetylation assay
Purified MeCP2e1-FLAG was incubated in deacetylation buffer (50 mM Hepes pH=7.9, 150 mM NaCl, 1mM DTT) in the presence of purified recombinant human SIRT1 (Sirtris Pharmaceuticals) plus 5 mM NAD+ (SIGMA), 1 mM trichostatin A (TSA), 50 mM EX527 (Tocris), 10 mM nicotinamide (NAM) for 1 h at 37°C. The reactions were resolved on SDS PAGE and analyzed by Western blotting.
In vitro acetylation assay
GST-MeCP2e1 was expressed in E. coli BL21. Recombinant proteins were lysed in lysis buffer (20 mM Tris-HCl pH=8, 0.3 mM EDTA, 20% Glycerol, 5mM DTT, 0.5 mM PMSF, 1% Triton X-100, 500 mM KCl) and purified by glutathione Sepharose 4B (Amersham). The purified protein was incubated in acetylation buffer (50 mM Tris-HCl pH=8, 50 mM AcetylCoA (SIGMA), 0.1 mM EDTA, 1mM DTT, 10% glycerol) plus 0.1 mg purified recombinant p300 catalytic domain (Active Motif). The reaction was incubated for 1 hr at 37 °C. Reactions were stopped by the addition of 2X Laemmli loading buffer, followed by SDS PAGE and Western blot analysis. Purified GST-MeCP2e1 was incubated in acetylation buffer (50 mM TrisHCl pH=8, 10% glycerol, 100 mM NaCl, 1 mM DTT, 0.2 mM EDTA, 0.2 mm PMSF, 1 mCi of 14CAcetyl-CoA) in the presence of 0.1 mg recombinant p300 (Active Motif). The reaction was stopped after 1 hr, and the samples were resolved by SDS-PAGE. Gels were stained by Coomassie blue, destained, dried and the level of acetyl MeCP2 was detected by autoradiography.
Antibodies and Western blot
Antibody against FLAG (M2) was from SIGMA, the anti-MeCP2 antibody was from Cell Signaling (MeCP2 D4F3 XPTM) and BDNF antibody from Santa Cruz (BDNF N20 sc-546). c-Myc was from Millipore (clone 9E10). Pan-acetyl lysine antibody (Cell Signaling #9441). The polyclonal acetyl-lysine 464 MeCP2e1 was generated by immunizing rabbits with KHL-conjugates of the peptide NH2-AEK(ac)YKHRGEGE (ABE28Millipore). Specificity of the antibody was validated both in vitro and in vivo by performing the appropriate controls, both in our laboratory and by Millipore/Merck. All Western blots were visualized using a chemiluminescence detection kit (Perkin-Elmer). At least three independent experiments were performed. Densitometry analysis of the film was performed using Adobe Photoshop.
Cell culture, cell extracts and immunoprecipitation.
Human embryonic kidney HEK-293 were maintained at 37°C and 5%CO2, in Dulbecco’s modified Eagle’s high glucose (Thermo Scientific) with antibiotics (penicillin and streptomycin) and 10% newborn calf serum (NCS). Cells were washed in phosphate-buffered saline (PBS) and lysed in RIPA buffer [50 mM Tris pH=8, 150 mM NaCl, 5 mM EDTA, 15 mM MgCl2, 1% NP40, 1X protease inhibitor cocktail (Roche), 1mM DTT, 1 mM trichostatin A (TSA), 10 mM NAM, 10 mM NaF, 1 mM PMSF]. Immunoprecipitation was performed by pre-clearing 500 mg-1mg of whole lysates with protein G-Agarose beads for two hours, and then by incubating with the appropriate amount of antibody or with anti FLAG-M2 Affinity gel at 4°C (SIGMA).
Plasmids
MeCP2e1-FLAG pCMV-TAG4 (STRATAGENE) was a kind gift of Dr. J.M. LaSalle. Mouse MeCP2e1 cDNA was cloned in a 6-myc/pCDNA3 plasmid. The point mutant MeCP2e1K464R was generated using the Agilent’s QuickChange site-directed mutagenesis kit. The truncation mutants of MeCP2 were generated by PCR amplification followed by cloning in 6-myc/pCDNA3 or pCMV-TAG4 (STRATAGENE). Stratagene QuickChange Site-Directed Mutagenesis Kit was used to generate the single point mutant K464R MeCP2e1.
Statistical analysis
Differences between two means were assessed with Student’s t-test. At least three independent experiments were performed (n=3). *p<0.05 was considered significant, **p<0.01 and ***p<0.001 were considered highly significant.
Additional material
Download Zip (898.9 KB)Acknowledgments
We are grateful to Li-Huei Tsai and Janine LaSalle for discussions, help and critical reading of the manuscript. We thank all the members of the Sassone-Corsi laboratory for helpful discussions. This work was supported by the National Institute of Health and Sirtris-GSK. L.Z. was in part supported by the American-Italian Cancer Foundation, New York.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Nan X, Campoy FJ, Bird A. MeCP2 is a transcriptional repressor with abundant binding sites in genomic chromatin. Cell 1997; 88:471 - 81; http://dx.doi.org/10.1016/S0092-8674(00)81887-5; PMID: 9038338
- Illingworth RS, Bird AP. CpG islands--‘a rough guide’. FEBS Lett 2009; 583:1713 - 20; http://dx.doi.org/10.1016/j.febslet.2009.04.012; PMID: 19376112
- Nan X, Ng HH, Johnson CA, Laherty CD, Turner BM, Eisenman RN, et al. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature 1998; 393:386 - 9; http://dx.doi.org/10.1038/30764; PMID: 9620804
- Chandler SP, Guschin D, Landsberger N, Wolffe AP. The methyl-CpG binding transcriptional repressor MeCP2 stably associates with nucleosomal DNA. Biochemistry 1999; 38:7008 - 18; http://dx.doi.org/10.1021/bi990224y; PMID: 10353812
- Guy J, Cheval H, Selfridge J, Bird A. The role of MeCP2 in the brain. Annu Rev Cell Dev Biol 2011; 27:631 - 52; http://dx.doi.org/10.1146/annurev-cellbio-092910-154121; PMID: 21721946
- Kriaucionis S, Bird A. The major form of MeCP2 has a novel N-terminus generated by alternative splicing. Nucleic Acids Res 2004; 32:1818 - 23; http://dx.doi.org/10.1093/nar/gkh349; PMID: 15034150
- Mnatzakanian GN, Lohi H, Munteanu I, Alfred SE, Yamada T, MacLeod PJ, et al. A previously unidentified MECP2 open reading frame defines a new protein isoform relevant to Rett syndrome. Nat Genet 2004; 36:339 - 41; http://dx.doi.org/10.1038/ng1327; PMID: 15034579
- Dragich JM, Kim YH, Arnold AP, Schanen NC. Differential distribution of the MeCP2 splice variants in the postnatal mouse brain. J Comp Neurol 2007; 501:526 - 42; http://dx.doi.org/10.1002/cne.21264; PMID: 17278130
- Amir RE, Van den Veyver IB, Wan M, Tran CQ, Francke U, Zoghbi HY. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat Genet 1999; 23:185 - 8; http://dx.doi.org/10.1038/13810; PMID: 10508514
- Kriaucionis S, Bird A. DNA methylation and Rett syndrome. Hum Mol Genet 2003; 12:Spec No 2 R221 - 7; http://dx.doi.org/10.1093/hmg/ddg286; PMID: 12928486
- Chahrour M, Zoghbi HY. The story of Rett syndrome: from clinic to neurobiology. Neuron 2007; 56:422 - 37; http://dx.doi.org/10.1016/j.neuron.2007.10.001; PMID: 17988628
- Martinowich K, Hattori D, Wu H, Fouse S, He F, Hu Y, et al. DNA methylation-related chromatin remodeling in activity-dependent BDNF gene regulation. Science 2003; 302:890 - 3; http://dx.doi.org/10.1126/science.1090842; PMID: 14593184
- Peddada S, Yasui DH, LaSalle JM. Inhibitors of differentiation (ID1, ID2, ID3 and ID4) genes are neuronal targets of MeCP2 that are elevated in Rett syndrome. Hum Mol Genet 2006; 15:2003 - 14; http://dx.doi.org/10.1093/hmg/ddl124; PMID: 16682435
- Swanberg SE, Nagarajan RP, Peddada S, Yasui DH, LaSalle JM. Reciprocal co-regulation of EGR2 and MECP2 is disrupted in Rett syndrome and autism. Hum Mol Genet 2009; 18:525 - 34; http://dx.doi.org/10.1093/hmg/ddn380; PMID: 19000991
- Nakagawa T, Guarente L. Sirtuins at a glance. J Cell Sci 2011; 124:833 - 8; http://dx.doi.org/10.1242/jcs.081067; PMID: 21378304
- Borrelli E, Nestler EJ, Allis CD, Sassone-Corsi P. Decoding the epigenetic language of neuronal plasticity. Neuron 2008; 60:961 - 74; http://dx.doi.org/10.1016/j.neuron.2008.10.012; PMID: 19109904
- Zocchi L, Sassone-Corsi P. Joining the dots: from chromatin remodeling to neuronal plasticity. Curr Opin Neurobiol 2010; 20:432 - 40; http://dx.doi.org/10.1016/j.conb.2010.04.005; PMID: 20471240
- Hisahara S, Chiba S, Matsumoto H, Tanno M, Yagi H, Shimohama S, et al. Histone deacetylase SIRT1 modulates neuronal differentiation by its nuclear translocation. Proc Natl Acad Sci U S A 2008; 105:15599 - 604; http://dx.doi.org/10.1073/pnas.0800612105; PMID: 18829436
- Prozorovski T, Schulze-Topphoff U, Glumm R, Baumgart J, Schröter F, Ninnemann O, et al. Sirt1 contributes critically to the redox-dependent fate of neural progenitors. Nat Cell Biol 2008; 10:385 - 94; http://dx.doi.org/10.1038/ncb1700; PMID: 18344989
- Kim D, Nguyen MD, Dobbin MM, Fischer A, Sananbenesi F, Rodgers JT, et al. SIRT1 deacetylase protects against neurodegeneration in models for Alzheimer’s disease and amyotrophic lateral sclerosis. EMBO J 2007; 26:3169 - 79; http://dx.doi.org/10.1038/sj.emboj.7601758; PMID: 17581637
- Qin W, Yang T, Ho L, Zhao Z, Wang J, Chen L, et al. Neuronal SIRT1 activation as a novel mechanism underlying the prevention of Alzheimer disease amyloid neuropathology by calorie restriction. J Biol Chem 2006; 281:21745 - 54; http://dx.doi.org/10.1074/jbc.M602909200; PMID: 16751189
- Donmez G, Wang D, Cohen DE, Guarente L. SIRT1 suppresses beta-amyloid production by activating the alpha-secretase gene ADAM10. Cell 2010; 142:320 - 32; http://dx.doi.org/10.1016/j.cell.2010.06.020; PMID: 20655472
- Min SW, Cho SH, Zhou Y, Schroeder S, Haroutunian V, Seeley WW, et al. Acetylation of tau inhibits its degradation and contributes to tauopathy. Neuron 2010; 67:953 - 66; http://dx.doi.org/10.1016/j.neuron.2010.08.044; PMID: 20869593
- Michán S, Li Y, Chou MM, Parrella E, Ge H, Long JM, et al. SIRT1 is essential for normal cognitive function and synaptic plasticity. J Neurosci 2010; 30:9695 - 707; http://dx.doi.org/10.1523/JNEUROSCI.0027-10.2010; PMID: 20660252
- Gao J, Wang WY, Mao YW, Gräff J, Guan JS, Pan L, et al. A novel pathway regulates memory and plasticity via SIRT1 and miR-134. Nature 2010; 466:1105 - 9; http://dx.doi.org/10.1038/nature09271; PMID: 20622856
- Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, et al. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science 2009; 325:834 - 40; http://dx.doi.org/10.1126/science.1175371; PMID: 19608861
- Dornan D, Shimizu H, Burch L, Smith AJ, Hupp TR. The proline repeat domain of p53 binds directly to the transcriptional coactivator p300 and allosterically controls DNA-dependent acetylation of p53. Mol Cell Biol 2003; 23:8846 - 61; http://dx.doi.org/10.1128/MCB.23.23.8846-8861.2003; PMID: 14612423
- Vaquero A, Scher M, Erdjument-Bromage H, Tempst P, Serrano L, Reinberg D. SIRT1 regulates the histone methyl-transferase SUV39H1 during heterochromatin formation. Nature 2007; 450:440 - 4; http://dx.doi.org/10.1038/nature06268; PMID: 18004385
- Zhou Z, Hong EJ, Cohen S, Zhao WN, Ho HY, Schmidt L, et al. Brain-specific phosphorylation of MeCP2 regulates activity-dependent Bdnf transcription, dendritic growth, and spine maturation. Neuron 2006; 52:255 - 69; http://dx.doi.org/10.1016/j.neuron.2006.09.037; PMID: 17046689
- Tao J, Hu K, Chang Q, Wu H, Sherman NE, Martinowich K, et al. Phosphorylation of MeCP2 at Serine 80 regulates its chromatin association and neurological function. Proc Natl Acad Sci U S A 2009; 106:4882 - 7; http://dx.doi.org/10.1073/pnas.0811648106; PMID: 19225110
- Guan JS, Haggarty SJ, Giacometti E, Dannenberg JH, Joseph N, Gao J, et al. HDAC2 negatively regulates memory formation and synaptic plasticity. Nature 2009; 459:55 - 60; http://dx.doi.org/10.1038/nature07925; PMID: 19424149
- Akhtar MW, Raingo J, Nelson ED, Montgomery RL, Olson EN, Kavalali ET, et al. Histone deacetylases 1 and 2 form a developmental switch that controls excitatory synapse maturation and function. J Neurosci 2009; 29:8288 - 97; http://dx.doi.org/10.1523/JNEUROSCI.0097-09.2009; PMID: 19553468