Abstract
The ability of ionizing radiation to initiate genomic instability has been harnessed in the clinic where the localized delivery of controlled doses of radiation is used to induce cell death in tumor cells. Though very effective as a therapy, tumor relapse can occur in vivo and its appearance has been attributed to the radio-resistance of cells with stem cell-like features. The molecular mechanisms underlying these phenomena are unclear but there is evidence suggesting an inverse correlation between radiation-induced genomic instability and global hypomethylation. To further investigate the relationship between DNA hypomethylation, radiosensitivity and genomic stability in stem-like cells we have studied mouse embryonic stem cells containing differing levels of DNA methylation due to the presence or absence of DNA methyltransferases. Unexpectedly, we found that global levels of methylation do not determine radiosensitivity. In particular, radiation-induced delayed genomic instability was observed at the Hprt gene locus only in wild-type cells. Furthermore, absence of Dnmt1 resulted in a 10-fold increase in de novo Hprt mutation rate, which was unaltered by radiation. Our data indicate that functional DNMTs are required for radiation-induced genomic instability, and that individual DNMTs play distinct roles in genome stability. We propose that DNMTS may contribute to the acquirement of radio-resistance in stem-like cells.
Introduction
Genome instability is a feature of cancer cells, manifesting in most cases as chromosomal instability (CIN), but in some endometrial and colorectal tumors as microsatellite or nucleotide-level instability (NIN).Citation1 This instability is an important part of oncogenesis and can be triggered by several environmental factors. Exposure to ionizing radiations is one such factor, which has been shown to promote malignancy in normal tissues.Citation2 Radiation-induced genomic instability is characterized in vitro by several endpoints including elevated levels of non-clonal mutations, chromosomal aberrations and cell death occurring within a clonal cell population at a delayed period after irradiation.Citation3 In vivo, the appearance of elevated levels of DNA damage, mutations and chromosomal instability can be detected in the un-irradiated progeny of irradiated animals.Citation2,Citation4
Although the mechanism by which radiation-induced delayed genomic instability is propagated is poorly understood,Citation5 several processes have been suggested as possible causes, including inflammatory type responses resulting in oxidative stressCitation6 and alterations in the balance of epigenetic factors such as DNA methylation.Citation7,Citation8 As a DNA-associated factor, DNA methylation is a promising candidate for the transmission and propagation of radiation-induced genomic instability.Citation7 In vivo studies have demonstrated the occurrence of radiation-induced reductions in DNA methylation in the radiation target tissue, in a genotype-, sex- and tissue-specific manner,Citation9,Citation10 while also suggesting that the least methylated tissues tend to be more radiosensitive.Citation9 In some instances, radiation-induced reductions in DNA methylation have been coupled with reduced expression levels of the DNA methyltransferases,Citation11 and significant accumulation of DNA strand breaks.Citation10
Aberrant hypomethylation, particularly when occurring at repetitive elements, including structural repeats and transposable sequences, is a common feature of cancer cells, and it is a major cause of genomic instability. Indeed, increased incidences of recombination and rearrangements at sub-telomeric repeats and pericentromeric classical satellite sequences,Citation12,Citation13 elevated levels of microsatellite instability,Citation14 reduced stability of triplet repeat sequencesCitation15 and activation of endogenous retroviral elements,Citation16 have all been shown to be associated with hypomethylation. Furthermore, consistent with increased genomic instability in hypomethylated cells is the observation that mouse embryonic stem cells (mESCs) lacking the maintenance DNA methyltransferase enzyme (DNMT1) display a 10-fold increase in mutation rate at Hprt and Tk gene loci.Citation17 Alongside global hypomethylation, both tumor cells and DNMT3A knockout cells show local hypermethylation particularly at CpG islands. The mechanism for the hypermethylation is unknown, but apparently not caused by overexpression of DNMTs.Citation18
In the clinic, controlled delivery of ionizing radiation to tumor tissues is used to cause local DNA damage and subsequent cell death making radiotherapy an extremely valuable and generally effective treatment for cancer.Citation19 However, several examples of radio-resistance and tumor relapse following radiotherapy have been reported, and it has been proposed that these may be due to the presence of a population of resistant cancer cells with stem cell features.Citation20 It is unknown whether cancer stem cells can survive irradiation and/or if the radiation can induce stem cell characteristics in some of the cancer cells. Regardless of the mechanism, presence of radio-resistant cancer cells with stem cell characteristics, i.e., the ability to initiate a tumor, become evident at delayed times after the initial irradiation. Cancer-initiating cells, or cancer stem cells, share several characteristics with embryonic stem cells, such as self-renewal capacities, the ability to form tumors, a common genetic program and similar DNA methylation profiles.Citation21,Citation22 To further investigate the relationship between DNA hypomethylation, radiosensitivity and delayed genomic instability in stem cells, we have used an established panel of five mESCs with differing levels of DNA methylation due to the presence (wild-type line J1)Citation23 or absence of maintenance (Dnmt1−/−)Citation24 or de novo (Dnmt3a−/−, Dnmt3b−/− and Dnmt3a3b–/–)Citation25 DNA methyltransferases. For our studies we chose a radiation dose of 3Gy X-rays. This is a leukemogenic dose of ionizing radiation,Citation26sufficient to induce hypomethylation of murine bone marrow at a delayed time point post irradiation.Citation9 Importantly for this study, 3Gy is a biologically relevant dose, close to the fractions sizes routinely used in radiotherapy, and we reasoned that use of higher doses would have reduced the biological relevance of this study.
Finally, analysis of cells immediately following radiation exposure was not included because the current study aimed to analyze instability at a delayed time point post radiation exposure rather than directly induced damage. The time points selected for the analyses were chosen to allow sufficient time for any changes in methylation or expression levels of genes to manifest. Time points were measured as population doublings (PDs) rather than days in culture to enable the comparison of the different mESC lines after the same number of cell divisions following irradiation with 0 or 3Gy X-ray (see Fig. S1 for the relationship between PDs and days in culture).
This study has allowed us to uncouple global DNA methylation and radiation-sensitivity while revealing unique roles played by individual DNA methyltransferases in genomic stability.
Results
Global levels of DNA methylation in mESCs remain constant following irradiation
Global levels of DNA methylation in the panel of mESCs were assessed by HPLC-UV and found to be consistent with reported data.Citation27,Citation28 Relative to wild-type mESCs, functional absence of DNMT1 substantially decreased genomic methylation, and while knockout of a single de novo methyltransferase caused a very modest reduction, functional absence of both DNMT3A and DNMT3B led to a decrease in cytosine methylation that declined further with increasing passages (). Nevertheless, none of the mESCs displayed significant differences in cytosine methylation levels as measured at 1 h, 24 h and 10 d post irradiation with 0Gy (sham treatment) or 3Gy X-ray (). There were no obvious abnormalities in the Dnmt knockout mESCs with respect to growth rate, morphology or cell cycle distribution in comparison to the wild-type cell line, either before or after irradiation (Figs. S1 and S2). The 1 and 24 h time points were included in this analysis because a previous study of radiation-induced alterations in methylation levels reported that changes can occur as early as 6 h post irradiation.Citation29 This time frame indicates that the causative mechanism is a driven response to radiation, rather than due to gradual loss of methylation through successive cell divisions. Thus, in analyzing whether radiation induced hypomethylation occurred in the 5 mESC lines, we included the early time points to assess whether any alterations observed were due to passive loss during cell division or by driven demethylation. The results, however, indicated that no alterations in methylation level occurred in the mESCs post irradiation. The inclusion of early time points in other experiments was thus deemed unnecessary as the main focus of this study were the delayed effects of radiation exposure.
Figure 1. Relationship between DNA methylation and irradiation. Genomic DNA cytosine methylation measured by HPLC-UV of un-irradiated mESC samples and mouse control tissues before (A) and after irradiation (B). Columns represent the average percentage of cytosine bases that were methylated. Error bars represent the standard error of the mean (SEM). Results were combined from multiple independent experiments. The number of replicate measurements for each sample is displayed above the error bar. (C) Dose-response curve of the ESC lines constructed using the long-term clonogenic assay after irradiation with 0–7Gy X-rays. Each data point represents the mean, and error bars represent the SEM, of at least 3 independent experiments. (D) Linear quadratic parameters α and β were calculated by fitting the data displayed in with a linear quadratic equation using SPSS and are shown in columns 3 and 5. Column 2 indicates the Plating Efficiency for each clone and column 4 and 6 the standard errors (SE) for α and β respectively. The survival fraction at 2Gy (SF2) inferred from the data are shown in the last column.
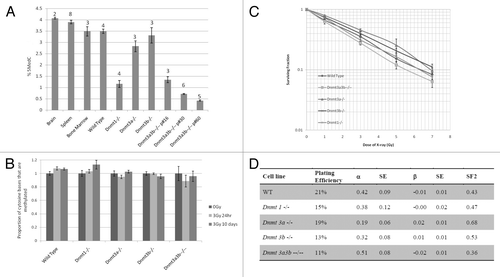
Radiosensitivity in mESCs is independent of global levels of DNA methylation
Long-term clonogenic survival assays on mESCs after irradiation with X-rays doses increasing from 0–7Gy revealed a dose-dependent decrease of the surviving fraction (SF) in all five lines (). Similarly to the data reported by other groups working with mESCs,Citation30 none of the cell lines showed any evidence of a shoulder, but instead displayed a near exponential survival curve (), as confirmed by the fact that the β values obtained by fitting a linear quadratic model to the data are all very close to zero (). Analysis of the survival curves showed no significant difference between the wild-type cell line and any of the hypomethylated Dnmt knockout cell lines. However, the Dnmt3a3b–/– cell line was significantly more radiosensitive than Dnmt3a−/− and Dnmt3b−/− (SF3Gy, p = 0.005 and p = 0.015, respectively). It must be noted that Dnmt3a3b–/– cells were analyzed at passage 20–25, when these cells have similar levels of DNA methylation to Dnmt1−/− cells (). As the two most hypomethylated cell lines (Dnmt3a3b–/– and Dnmt1−/−) display similar clonogenic survival responses to radiation as the wild-type cell line, it seems that hypomethylation alone is not sufficient to increase radiosensitivity. Thus, the DNMT3 family of enzymes may contribute to the determination of radiosensitivity in mESCs, and the absence of a single de novo methyltransferase, particularly DNMT3A, may result in slight radioresistance.
Delayed effects of radiation as measured by DNA damage or oxidative stress are comparable in wild-type and hypomethylated mESCs
Radiation induced delayed genomic instability is characterized by the persistent expression of elevated levels of non-clonal mutations or aberrations at a delayed period after irradiation, which are detected in the un-irradiated progeny of the irradiated cellsCitation31 or animalsCitation2 several generations after irradiation.Citation4 The persistent presence of elevated levels of DNA damage, manifesting as strand breaks and oxidative damage in the un-irradiated progeny of irradiated cells, is an accepted indicator of ongoing genomic instability, and the Comet assay has successfully been used to detect such ongoing instability in multiple previous studies.Citation2,Citation32 We have used the Comet assay as another measure of delayed instability and we thus looked at late time points to analyze those cells surviving, and “recovered from,” the effect of irradiation, i.e., the un-irradiated progeny of the irradiated cells.
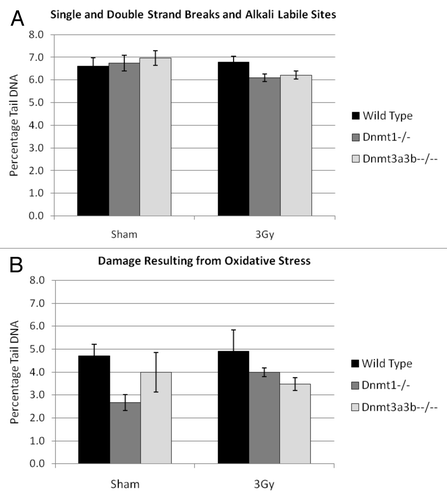
Average levels of single-strand breaks (SSBs), double-strand breaks (DSBs) and alkali-labile sites (ALS) were determined in wild-type, Dnmt1−/− and Dnmt3a3b–/– mESCs at 23–25 population doublings (PDs) after treatment with 0 or 3Gy X-irradiation using the alkaline comet assay. The average level of endogenous and/or induced DNA damage was found to be very similar between all of the cell lines and treatment conditions (p > 0.07) (). The Fpg-modified comet assay was then used to assess the level of oxidative damage in wild-type, Dnmt1−/− and Dnmt3a3b–/– mESCs. After correction for damage levels observed in the non- enzyme treatment controls, there was no significant difference in the average level of FPG-sensitive lesions between any of the cell lines (p > 0.07), or between the 3Gy and 0Gy (sham) treatment conditions (p > 0.06), indicating no differences in the level of damage induced by long-term oxidative stress ().
Figure 2. Analysis of single strand, double strand and alkali labile site DNA damage (A) or damage resulting from oxidative stress (B) in wild-type, Dnmt1−/−. Dnmt3a3b–/– mESC lines, 23–25PDs post 3Gy X-irradiation or sham treatment as measured by percentage of tail DNA. Results are expressed as an average of 3 independent experiments, each comprising 200 separate cell measurements. Error bars represent the SEM of the averages of the 3 independent experiments.
Radiation-induced delayed chromosomal instability is independent of DNA methylation levels
Next, the frequencies of cytogenetic aberrations were assessed in wild-type, Dnmt1−/− and Dnmt3a3b–/– mESC lines and the structural cytogenetic aberrations observed were grouped according to defined classificationsCitation33 at 10–14 () and 23–25 () population doublings post irradiation with 0Gy or 3Gy X-rays (see for details). A mid-time point of 10–14 PDs was used in the cytogenetic analyses, in order to see whether levels of aberrations varied over time post irradiation and the results of cells examined post exposure were always compared with the results for un-exposed control cells for the same time point. There were no significant differences in the frequency of total structural aberrations or of any of the individual aberration types between any of the cell lines, treatment groups or time points, as calculated using the chi-square test (p > 0.14 and p > 0.11, respectively). The average number of aberrations per metaphase was also not significantly different between any of the cell lines, treatment conditions or time points (p > 0.12, data not shown).
Figure 3. Classifications of structural cytogenetic aberrations as defined by Savage.Citation33 The results are displayed as the number of each class of aberration observed per 100 metaphases. Graph (A) displays the results 10–14 PDs post treatment; Graph (B) displays the results 23–25 PDs post treatment. (C) Mitotic spreads of Giemsa-stained chromosomes from the Dnmt1−/− ESC line 23–25 PDs after 0Gy treatment, showing a chromosome with very long satellite arms. Immediately after exposure to 3Gy X-rays or sham treatment, cells were seeded into the clonogenic assay. Surviving colonies were counted 12–14 d after seeding. (D) Dicentric, tri-centric and tetra-centric chromosome-type aberrations observed in Giemsa-stained metaphase spreads of Dnmt1−/− and Dnmt3a3b–/– mESCs 23–25PDs post 0Gy (sham) or 3Gy treatment.
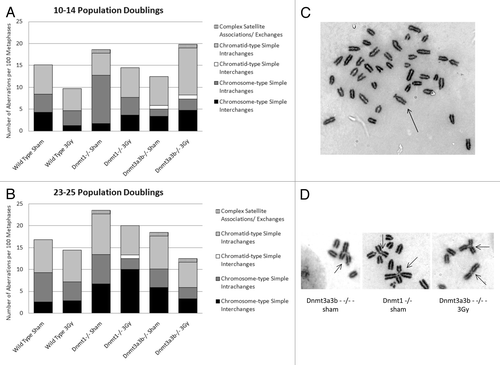
Table 1. Absolute numbers of each cytogenetic aberration type observed at 10-14 PD and 23-25 PD post treatment with 0Gy or 3Gy X-rays
A high ratio of chromatid:chromosome type aberrations is accepted as indicative of ongoing radiation-induced chromosomal instability in cultures of cells.Citation33,Citation34 In this study, the chromatid:chromosome type aberrations ratio observed in the wild-type cell line was comparable at both time points (). In addition, this ratio did not differ significantly between the 3Gy and sham treatment groups for any of the mESC lines at both time points.
Possible relationship between methylation levels and aberrations involving the satellite arms
An aberration involving the appearance of abnormally long satellite arms was observed almost exclusively in the Dnmt1−/− cell line at the 23–25 PD time point (). It occurred once in the 3Gy-treated wild-type cells, 8 times in the 3Gy-treated Dnmt1−/− cells, and 5 times in the sham-treated Dnmt1−/− cells (see for frequency of abnormalities). As it could not be determined whether these aberrations were clonal, a conservative approach was taken and multiple appearances within the same cell population were counted as a single aberration. In addition, several complex exchanges involving the satellite arms of 3 or more chromosomes were observed solely in the hypomethylated Dnmt1−/− and Dnmt3a3b–/– mESC lines, at both time points and irrespective of the treatment condition (). Assuming that the long satellite arm aberration was clonal, the difference in total frequency of aberrations involving associations or exchanges of the satellite arms in comparison to wild-type cells was not statistically significant for Dnmt1−/− mESCs (p = 0.17), while it displayed a significant increase in Dnmt3a3b–/– cells (p = 0.006).
DNMTs are required for radiation-induced delayed genomic instability
As we did not detect any highly significant differences in delayed instability cytogenetically between the mESC lines, we performed the analysis at a higher resolution level by comparing mutation rates at a single gene locus, Hprt. Locus specific delayed genomic instability was assessed in mESCs cultured in the presence of 6TG and the mutation rate at the Hprt locus was measured 23–25 population doublings after irradiation with 0Gy and 3Gy X-rays (sham and 3Gy in , respectively). This permitted analysis of the de novo mutation rate of each cell line in addition to the radiation-induced delayed mutation rate (see for details of selection). The appearance of individual colonies in the 6 TG selection dishes revealed that no founder mutations were present in any of the clonal populations.
Figure 4. (A) Hprt gene mutation rates 23–25 population doublings after 3Gy X-irradiation or 0Gy (sham) treatment. Columns represent the mutation rate, calculated from 19–25 individual clonal expansions using Luria-Delbrück fluctuation analysis and corrected for plating efficiency. Error bars represent the 95% confidence intervals. Dnmt3a3b–/– mESCs were p#20–26 at the time of selection for mutants. (B) Spectrum of functional Hprt gene mutations observed in mESC lines 23–25 population doublings after 3Gy X-irradiation or 0Gy (sham) treatment. (C) Tables detailing the mutations identified by exonic PCR that were used to construct the pie charts in (B). PCRs were performed using primer pairs designed to amplify each exon (1–9) of the Hprt gene and exon 2 of the unlinked K-ras gene as a control. When a mutation was observed in multiple colonies arising in the same clonal population it was counted only once. ‘Other mutations’ include mutations that could not be detected using the PCR-based method, such as point mutations, frame shift mutations and epigenetic alterations.
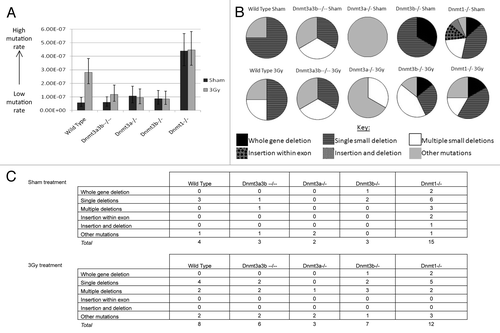
Table 2. Details of the selections carried out to determine the mutation rate at the Hprt gene for each cell line and treatment condition.
Mutation rates were calculated using Luria-Delbrück fluctuation analysis.Citation35 The de novo mutation rates of the Dnmt3a−/−, Dnmt3b−/− and Dnmt3a3b–/– mESCs were not significantly different from the wild-type mESC line (p > 0.2) (). Therefore, global hypomethylation does not appear to influence mutation rate at the Hprt locus in mESCs. However, in agreement with previous reports,Citation17 the Dnmt1−/− mESCs displayed a 10-fold elevated de novo mutation rate in comparison to the wild-type mESCs (p = 2.7x10−6), indicating the importance of DNMT1 for genomic stability in mESCs (). Unexpectedly, the de novo mutation rate of Dnmt1−/− mESCs was significantly higher than that of Dnmt3a3b–/– mESCs (p = 3.2x10−6). Dnmt3a3b–/– mESCs were at passage 20–26 at the time of seeding into selective media and therefore had a very similar low global methylation level to Dnmt1−/− mESCs (). Thus, it appears that the elevated mutation rate detected in the Dnmt1−/− mESC line is not a result of global hypomethylation but a result of the absence of a property or function specific to DNMT1 or of loss of methylation at genomic regions specifically targeted by DNMT1.
Wild-type mESCs displayed a significantly elevated mutation rate in the 3Gy X-irradiated clones in comparison to the sham treated control clones at 23–25 population doublings post treatment (p = 1.6x10−4). This indicates the occurrence of radiation-induced delayed genomic instability. However, none of the Dnmt knockout mESCs showed a significantly elevated mutation rate in the 3Gy X-irradiated clones in comparison to their corresponding sham treated controls (p > 0.4) (). Thus, Dnmt knockout mESCs showed no evidence of radiation-induced delayed genomic instability. These results indicate that absence of the DNA methyltransferase enzymes, or disruption of the normal methylation pattern, may potentially inhibit the mechanism behind radiation-induced delayed genomic instability at the nucleotide level.
Increased retrotransposition in Dnmt1−/− mES cells
To investigate the mechanisms that might be responsible for the differing mutation rates between the ESC lines, the types of mutations present in the Hprt gene were characterized using exonic PCRs on DNA from 6-TG resistant clones from each cell line and treatment condition. The spectra of mutations detected in each cell line after sham or 3Gy treatment are displayed in ; displays the numbers of each mutation type identified in sham treated and 3Gy irradiated cells. The details of mutants obtained and analyzed by PCR are summarized in Table S1. Multiple deletions of non-contiguous exons were observed in sham treated clones from the two most hypomethylated mESC lines (Dnmt1−/− and Dnmt3a3b–/–) and in clones derived from all of the mESC lines after irradiation with 3Gy X-ray (Fig. S3). Hypomethylated Dnmt1−/− mESCs, which had the highest de novo mutation rate, also displayed the widest variety of mutation types, indicating that several different mutational processes may occur in this cell line. Dnmt1−/− cells were the only line to demonstrate the occurrence of active retrotransposition, indicated by insertions involving two different subfamilies of non-autonomous SINE retrotransposons (B2 and B1/Alu) 5′ of exon 3 and exon 6 respectively (Fig.S4).
It is possible that active retrotransposition was not observed in the other lines due to their lower spontaneous mutation rates. However, the high de novo mutation rate of Dnmt1−/− mESCs was further demonstrated by the occurrence of second mutation events during expansion of the clones. For example, one of the many colonies carrying the B1 SINE insertion was also found to have a deletion of exon 4, which must have arisen at a delayed period after the insertion. As SINEs require the LINE-1 encoded ORF2 protein for their retrotransposition,Citation36 LINE-1 methylation status was tested in these cells. Although all the mESCs are hypomethylated relative to control tissues, Dnmt1−/− and Dnmt3a3b–/– LINE-1s were slightly less methylated than wild-type mESCs (Fig. S5), suggesting that LINE-1 hypomethylation per se is not sufficient to promote retrotransposition as Dnmt3a/3b–/–mESCs do not show such an effect.
Discussion
The work described in this study was aimed to examine the relationship between DNA methylation, radiosensitivity and delayed genomic instability in mouse embryonic stem cells. Rather than using inhibitors of DNA methyltransferases such as 5-azacytidine, which have a broad range of effects, we addressed the roles of maintenance (DNMT1) and de novo (DNMT3A and DNMT3B) DNA methyltransferases using a panel of mECS that is well established for the studies of activities of individual DNMTs. This panel consists of five mESC lines including the wild-type line J1Citation23 and four of its derivatives (Dnmt1−/−, Dnmt3a−/−, Dnmt3b−/− and Dnmt3a3b–/–) in which the DNA methyltransferases were inactivated by targeted deletions within their catalytic domains.Citation24,Citation25
There are reports describing alterations in methylation levels post 3Gy X-irradiation in somatic tissues in vivo,Citation9,Citation29 and a correlation between global DNA hypomethylation and increased radiosensitivity typical of somatic cell lines.Citation37,Citation38 However, radiation-induced methylation was observed to vary in a strain-, tissue- and sex-specific manner. We have now shown that in mESCs, exposure to 3Gy X-irradiation does not lead to changes in DNA methylation, and that radiosensitivity is independent of DNA methylation levels. These findings may reflect unique properties of mESCs in response to radiation that distinguish them from somatic cells, though further studies will be required to confirm this. Given the cell- and tissue-type specificity of these phenomena,Citation39 our finding may represent a property of mESCs reflecting their hypomethylation in comparison to most somatic cells. Consistent with previous observations, all five mESC lines displayed nearly exponential survival curves,Citation30 which may reflect their increased propensity to undergo apoptosis in response to DNA damage.Citation40 Yet, the de novo methyltransferases, DNMT3A and DNMT3B, may play a role in modulating sensitivity to X-rays in mESCs, as their absence seems to have a modest radioprotective effect.
Although levels of radiation-induced delayed chromosomal instability were comparable in all the mESCs lines before and after irradiation, aberrations and multicentric exchanges/associations involving the satellite arms of chromosomes were present at an increased frequency in hypomethylated mESCs (Dnmt1−/− and Dnmt3a3b–/–) compared with wild-type cells (). It is possible that increased frequencies of rearrangements or homologous recombination at the major and minor satellite repeats occur due to hypomethylation. Repetitive sequences, such as retroviral elements and satellite repeats, contain the majority of the methyl-cytosine in mammalian cells,Citation41 can be hotspots for recombinationCitation42 and are hypomethylated in patients carrying DNMT1 mutations.Citation43 Gonzalo et al.Citation12 demonstrated that Dnmt1−/− and Dnmt3a3b–/– mESCs display greater heterogeneity of telomere length and significantly increased frequencies of telomere sister chromatid exchange (T-SCE). Interestingly, exchanges between hypomethylated satellite sequences, resulting in the formation of multi-radial chromosomes, are frequently observed in lymphocytes from patients with ICF syndrome, which carry mutations in DNMT3B.Citation13 Further work is required to determine whether the frequency of recombination throughout the genome is elevated in the hypomethylated Dnmt1−/− and Dnmt3a3b–/– mESCs.
Sham treated Dnmt1−/− mESCs displayed an approximately 10-fold elevation of de novo mutation rate at the X-linked Hprt gene locus in comparison to that of wild-type mESCs, while all the other cell lines did not differ from wild-type cells. A previous report has described an increase in mutation rates in Dnmt1−/− cells; however, only Dnmt1−/− cells were investigated, without any irradiation treatment, and it was concluded that the increased mutation rate was a result of DNA hypomethylation.Citation17 We have now demonstrated that this elevated mutation rate is not simply a result of global hypomethylation. If that were the case, a comparable increase in de novo mutation rate would be expected for the similarly hypomethylated Dnmt3a3b–/– mESC line. Instead, the mutation rate in this cell line does not differ from that of wild-type cells. Rather, the fact that the increase in mutation rate is specific to Dnmt1−/− cells suggests that it is caused by the absence of methylation in regions specifically targeted by DNMT1 or of other properties or functions specific to DNMT1. Such functions may include the various roles played by DNMT1 in DNA replication and repair. DNMT1, but not DNMT3A or DNMT3B, is recruited by PCNA to the replication fork during S phase of the cell cycle, and also to sites of DNA repair.Citation44 Furthermore, DNMT1 is thought to play a role in strand discrimination during mismatch repair (MMR).Citation45 Thus, it is possible that absence of functional DNMT1 or the lack of methylation of genes and regions targeted by DNMT1 may hinder the efficient functioning of DNA repair, causing an increase in mutation rate. In addition, mobilization of retrotransposons in Dnmt1−/− cells may contribute to the overall increase in mutation rate, an observation consistent with recent evidence that degradation of DNMT1 following exposure to benza(a)pyrene leads to L1 reactivation.Citation46 A hypothesis that remains to be explored is whether there is also an elevated mutation rate in patients carrying DNMT1 mutations that lead to late onset, neurological conditions.Citation43,Citation47
Non-contiguous deletions at the Hprt locus were seen in the most hypomethylated lines (Dnmt1−/− and Dnmt3a3b–/–) before irradiation, and in all lines post irradiation. We propose that their appearance may reflect increased homologous recombination (HR). Ionizing radiation is known to induce a persistent increase in homologous recombination,Citation4 which can be detected as sister chromatid exchanges (SCEs).Citation48 Furthermore, Rugo et al. (2011) found that naive mESCs exhibited a significant increase in the frequency of SCEs when they shared media with descendants of irradiated mESCs, and that SCEs were increased in cells that shared media with un-irradiated triple DNMT knockout mESCs, compared with cells that shared media with un-irradiated wild-type mESCs.Citation32 Further studies will be required for the detection of SCEs in our system.
In contrast to what we observed in wild-type mESCs, none of the Dnmt knockout cells displayed radiation-induced delayed genomic instability, a direct demonstration that specific DNA methylation patterns and/or the DNA methyltransferases are required for this phenomenon to occur. These findings are largely congruent with recently published data obtained studying the bystander effect,Citation32 and indicate that active DNMTs are required for persistent radiation-induced genomic instability. It has been proposed that alterations in DNMT activity or the DNA methylation profile could potentially alter the secretion profiles of cells for factors such as TNF-α, NO and TGF-β, which impact genomic stability.Citation32 The inheritance of such altered methylation patterns could potentially result in propagation of the instability phenotype through successive generations (as seen in un-irradiated Dnmt1−/− mESCs), or lack of appearance of the instability phenotype after radiation exposure (as seen in all Dnmt knockout mESCs exposed to 3Gy X-rays). Further investigation of the expression levels of such factors in the mESCs would be necessary to determine which, if any, of these possible scenarios is true. The data point to an apparent paradoxical nature of DNMT1 loss, which can both promote (un-irradiated Dnmt1−/− mESCs) and prevent (irradiated Dnmt1−/− mESCs) delayed genomic instability. However, the lack of increase in genomic instability levels in Dnmt1−/− cells following irradiation may be a reflection of the already high mutation rates in un-irradiated cells. Future experiments, including the rescue of knockout cells with the respective Dnmts and treatments with demethylating agents and/or specific DNMT inhibitors, will be required to investigate whether absence of functional DNMTs or disruption of the normal methylation pattern inhibit the causative mechanism behind radiation-induced delayed genomic instability or the appearance of the effects.
In summary, we have provided experimental evidence suggesting that radiosensitivity or delayed genomic instability in mESCs is independent of global levels of genomic DNA hypomethylation, but that individual DNMTs play distinct roles in these responses, whereby the de novo DNA methyltransferases may influence radiosensitivity and are required for radiation-induced genomic instability, and functional DNMT1 is required to maintain the stability of genes such as Hprt. As mouse embryonic stem cells share several characteristics with cancer stem cells, our data support the hypothesis that radioresistance and genomic instability in cancer may in some instances be attributed to changes in activity of DNMTs (e.g., ref. Citation49). Analysis of these factors in normal and tumor tissues will further our understanding of the conditions/processes required for the manifestation of radiation induced genomic instability, which may eventually lead to oncogenesis and/or radioresistance. At the same time, this experimental system will allow a further characterization of the mechanisms by which lack of DNMT activities contributes to these phenomena and the identification of methods to modulate radiosensitivity in tumors and normal tissues, thus contributing to the design and optimization of future therapies.
Materials and Methods
Sequences of oligonucleotides can be found in Tables S2 and S3.
mESCs culture and irradiation
mESCs cell lines were maintained on gelatinized plates in DMEM high glucose (Invitrogen) with ESGRO (Millipore) as previously described.Citation50 mESCs were irradiated with 1-, 3-, 5-, 7- or 10-Gy of X-rays delivered at 0.5Gy min−1 (250 kV constant potential, HLV 1.5mm Cu), using a Pantak industrial X-ray machine, and media changed immediately after irradiation.
DNA methylation analysis by HPLC-UV
Genomic DNA was extracted using QIAamp DNA Mini Kit (Qiagen) following treatment of cell pellets with RNase A (Roche Diagnostics, 10mg/ml) and RNase T1 (Roche Diagnostics, 100,000U/ml). Global levels of DNA cytosine methylation were characterized using high performance liquid chromatography with UV detection (HPLC-UV) as previously described.Citation39
Clonogenic assays
Clonogenic assays were performed as previously describedCitation51 by seeding 103, 2x103, 4x103 and 5x103 cells for 0, 1, 3, 5, and 7Gy treated mESCs respectively. Plating efficiency (PE) was assessed by the proportion of cells seeded into the 0Gy well that survived to produce colonies. Statistical analysis was performed using a two-tailed T-test.
Comet assays
Alkaline Comet Assays were based on previously described protocolsCitation52 and performed 23–25 population doublings post 3Gy X-irradiation or sham treatment. Images were analyzed with the Komet Analysis software version 5.5 (Andor Bioimaging). 200 cells were analyzed per sample per experiment. Percentages of the cellular DNA within each comet tail were calculated (% tail DNA), and results expressed as the mean and standard error of the mean (SEM) of three independent experiments. Control slides prepared using wild-type mESCs irradiated with 0, 5 and 10Gy X-Rays immediately prior to the Comet assay were included to ensure consistency between experiments.
The relative levels of oxidative purine base damage were monitored using the human formamidopyrimidine DNA glycosylase (fpg) comet assay (fpg comet) as described previously for the human 8-oxoguanine DNA glycosylase 1 (hOGG1) comet assay (hOGG1 comet)Citation53 with the following modifications: after lysis the slides were washed once with distilled water and immersed in three changes of enzyme digestion buffer [40 mM HEPES, 0.1 M KCl, 0.5 mM EDTA and 0.2 mg/ml bovine serum albumin (pH 8.0)], for 5 min each time, on ice. Fpg (Sigma-Aldrich) was added to the gel (50 μl/gel) at 1/500 dilution; gels were covered with a coverslip and incubated in a humidified chamber at 37°C for 30 min; the coverslips were removed and the slides were placed in a horizontal electrophoresis tank. From this step onwards, the assay was performed as described.Citation54 DNA damage was expressed as the percentage of DNA in the comet tails.
Cytogenetic aberrations
Cells were treated with 0.03μg/ml KaryoMAX colcemid solution (GIBCO) eight hours after irradiation with 0 or 3Gy X-ray and slides prepared for cytogenetic analysis as previously described.Citation55 Scoring was performed with bright field function and a green filter on a Carl Zeiss Microscope (Axioskop 2). Pictures were taken using Ikaros (MetaSystems) or Smart Capture X (Digital Scientific UK) imaging software. P values were calculated using the chi-square test.
Hprt mutation analysis
Analysis of mutation rate at Hprt on 0Gy or 3Gy X-rays treated cells was performed according to the 6-thioguanine (6TG) selection protocol as described.Citation17 Mutation rates were calculated using the maximum likelihood method of Luria-Delbrück fluctuation analysis implemented in Salvador v2.3.Citation35 Exonic mutations were studied as previously described.Citation56
| Abbreviations: | ||
| DNMT | = | DNA methyltransferase |
| mESCs | = | mouse embryonic stem cells |
| HPLC-UV | = | high performance liquid chromatography with ultraviolet detection |
| 6-TG | = | 6-thioguanine |
| SINE | = | short interspersed nuclear element |
| LINE | = | long interspersed nuclear element |
| ICF | = | immunodeficiency, centromere instability and facial anomalies |
| PCNA | = | proliferating cell nuclear antigen |
Additional material
Download Zip (1 MB)Acknowledgments
The intellectual basis to this project was in a large part due to Dr. Mark Plumb, and we dedicate this paper to him. We would like to thank En Li and Taiping Chen for their kind gift of the mESC lines, and Munira Kadhim and Debbie Bowler for cytogenetic training. We are grateful for technical assistance from George Giotopoulos and Rohan Machhar. Finally, most felt thanks go to Yuri Dubrova for continued support, constructive discussions and critical reading of the manuscript. Funding for consumables came from NOTE and from the BBSRC for the PhD studentship to CAA. CT was supported by a Royal Society Dorothy Hodgkin Research Fellowship.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Financial Disclosures
Research was supported by NOTE, CAA was supported by a PhD studentship from BBSRC and CT by a Royal Society Dorothy Hodgkin Research Fellowship. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- Lengauer C, Kinzler KW, Vogelstein B. Genetic instabilities in human cancers. Nature 1998; 396:643 - 9; http://dx.doi.org/10.1038/25292; PMID: 9872311
- Barber RC, Hickenbotham P, Hatch T, Kelly D, Topchiy N, Almeida GM, et al. Radiation-induced transgenerational alterations in genome stability and DNA damage. Oncogene 2006; 25:7336 - 42; http://dx.doi.org/10.1038/sj.onc.1209723; PMID: 16751800
- Little JB, Nagasawa H, Pfenning T, Vetrovs H. Radiation-induced genomic instability: delayed mutagenic and cytogenetic effects of X rays and alpha particles. Radiat Res 1997; 148:299 - 307; http://dx.doi.org/10.2307/3579514; PMID: 9339945
- Huang L, Grim S, Smith LE, Kim PM, Nickoloff JA, Goloubeva OG, et al. Ionizing radiation induces delayed hyperrecombination in Mammalian cells. Mol Cell Biol 2004; 24:5060 - 8; http://dx.doi.org/10.1128/MCB.24.11.5060-5068.2004; PMID: 15143196
- Wright EG. Manifestations and mechanisms of non-targeted effects of ionizing radiation. Mutat Res 2010; 687:28 - 33; http://dx.doi.org/10.1016/j.mrfmmm.2010.01.014; PMID: 20080112
- Lorimore SA, Chrystal JA, Robinson JI, Coates PJ, Wright EG. Chromosomal instability in unirradiated hemaopoietic cells induced by macrophages exposed in vivo to ionizing radiation. Cancer Res 2008; 68:8122 - 6; http://dx.doi.org/10.1158/0008-5472.CAN-08-0698; PMID: 18829571
- Dubrova YE. Radiation-induced transgenerational instability. Oncogene 2003; 22:7087 - 93; http://dx.doi.org/10.1038/sj.onc.1206993; PMID: 14557814
- Kaup S, Grandjean V, Mukherjee R, Kapoor A, Keyes E, Seymour CB, et al. Radiation-induced genomic instability is associated with DNA methylation changes in cultured human keratinocytes. Mutat Res 2006; 597:87 - 97; http://dx.doi.org/10.1016/j.mrfmmm.2005.06.032; PMID: 16417911
- Giotopoulos G, McCormick C, Cole C, Zanker A, Jawad M, Brown R, et al. DNA methylation during mouse hemopoietic differentiation and radiation-induced leukemia. Exp Hematol 2006; 34:1462 - 70; http://dx.doi.org/10.1016/j.exphem.2006.06.008; PMID: 17046565
- Koturbash I, Pogribny I, Kovalchuk O. Stable loss of global DNA methylation in the radiation-target tissue--a possible mechanism contributing to radiation carcinogenesis?. Biochem Biophys Res Commun 2005; 337:526 - 33; http://dx.doi.org/10.1016/j.bbrc.2005.09.084; PMID: 16202973
- Raiche J, Rodriguez-Juarez R, Pogribny I, Kovalchuk O. Sex- and tissue-specific expression of maintenance and de novo DNA methyltransferases upon low dose X-irradiation in mice. Biochem Biophys Res Commun 2004; 325:39 - 47; http://dx.doi.org/10.1016/j.bbrc.2004.10.002; PMID: 15522198
- Gonzalo S, Jaco I, Fraga MF, Chen T, Li E, Esteller M, et al. DNA methyltransferases control telomere length and telomere recombination in mammalian cells. Nat Cell Biol 2006; 8:416 - 24; http://dx.doi.org/10.1038/ncb1386; PMID: 16565708
- Tuck-Muller CM, Narayan A, Tsien F, Smeets DF, Sawyer J, Fiala ES, et al. DNA hypomethylation and unusual chromosome instability in cell lines from ICF syndrome patients. Cytogenet Cell Genet 2000; 89:121 - 8; http://dx.doi.org/10.1159/000015590; PMID: 10894953
- Kim M, Trinh BN, Long TI, Oghamian S, Laird PW. Dnmt1 deficiency leads to enhanced microsatellite instability in mouse embryonic stem cells. Nucleic Acids Res 2004; 32:5742 - 9; http://dx.doi.org/10.1093/nar/gkh912; PMID: 15509869
- Dion V, Lin Y, Hubert L Jr., Waterland RA, Wilson JH. Dnmt1 deficiency promotes CAG repeat expansion in the mouse germline. Hum Mol Genet 2008; 17:1306 - 17; http://dx.doi.org/10.1093/hmg/ddn019; PMID: 18252747
- Iskow RC, McCabe MT, Mills RE, Torene S, Pittard WS, Neuwald AF, et al. Natural mutagenesis of human genomes by endogenous retrotransposons. Cell 2010; 141:1253 - 61; http://dx.doi.org/10.1016/j.cell.2010.05.020; PMID: 20603005
- Chen RZ, Pettersson U, Beard C, Jackson-Grusby L, Jaenisch R. DNA hypomethylation leads to elevated mutation rates. Nature 1998; 395:89 - 93; http://dx.doi.org/10.1038/25779; PMID: 9738504
- Challen GA, Sun D, Jeong M, Luo M, Jelinek J, Berg JS, et al. Dnmt3a is essential for hematopoietic stem cell differentiation. Nat Genet 2012; 44:23 - 31; http://dx.doi.org/10.1038/ng.1009; PMID: 22138693
- Joiner MC, Van Der Kogel AJ, Steel GG. Introduction: the significance of radiobiology and radiotherapy for cancer treatment. In:MJAvd Kogel. Basic Clinical Radiobiology. 4th.Edward Arnold, UK, 2009:1.
- Woodward WA, Bristow RG. Radiosensitivity of cancer-initiating cells and normal stem cells (or what the Heisenberg uncertainly principle has to do with biology). Semin Radiat Oncol 2009; 19:87 - 95; http://dx.doi.org/10.1016/j.semradonc.2008.11.003; PMID: 19249646
- Calvanese V, Horrillo A, Hmadcha A, Suarez-Alvarez B, Fernandez AF, Lara E, et al. Cancer genes hypermethylated in human embryonic stem cells. PLoS One 2008; 3:e3294; http://dx.doi.org/10.1371/journal.pone.0003294; PMID: 18820729
- Wong DJ, Liu H, Ridky TW, Cassarino D, Segal E, Chang HY. Module map of stem cell genes guides creation of epithelial cancer stem cells. Cell Stem Cell 2008; 2:333 - 44; http://dx.doi.org/10.1016/j.stem.2008.02.009; PMID: 18397753
- Li E, Bestor TH, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell 1992; b 69:915 - 26; http://dx.doi.org/10.1016/0092-8674(92)90611-F; PMID: 1606615
- Lei H, Oh SP, Okano M, Jüttermann R, Goss KA, Jaenisch R, et al. De novo DNA cytosine methyltransferase activities in mouse embryonic stem cells. Development 1996; 122:3195 - 205; PMID: 8898232
- Okano M, Xie S, Li E. Cloning and characterization of a family of novel mammalian DNA (cytosine-5) methyltransferases. Nat Genet 1998; 19:219 - 20; http://dx.doi.org/10.1038/890; PMID: 9662389
- Wakeford R. The cancer epidemiology of radiation. Oncogene 2004; 23:6404 - 28; http://dx.doi.org/10.1038/sj.onc.1207896; PMID: 15322514
- Chen T, Ueda Y, Dodge JE, Wang Z, Li E. Establishment and maintenance of genomic methylation patterns in mouse embryonic stem cells by Dnmt3a and Dnmt3b. Mol Cell Biol 2003; 23:5594 - 605; http://dx.doi.org/10.1128/MCB.23.16.5594-5605.2003; PMID: 12897133
- Jackson M, Krassowska A, Gilbert N, Chevassut T, Forrester L, Ansell J, et al. Severe global DNA hypomethylation blocks differentiation and induces histone hyperacetylation in embryonic stem cells. Mol Cell Biol 2004; 24:8862 - 71; http://dx.doi.org/10.1128/MCB.24.20.8862-8871.2004; PMID: 15456861
- Pogribny I, Raiche J, Slovack M, Kovalchuk O. Dose-dependence, sex- and tissue-specificity, and persistence of radiation-induced genomic DNA methylation changes. Biochem Biophys Res Commun 2004; 320:1253 - 61; http://dx.doi.org/10.1016/j.bbrc.2004.06.081; PMID: 15249225
- Sioftanos G, Ismail A, Föhse L, Shanley S, Worku M, Short SC. BRCA1 and BRCA2 heterozygosity in embryonic stem cells reduces radiation-induced Rad51 focus formation but is not associated with radiosensitivity. Int J Radiat Biol 2010; 86:1095 - 105; http://dx.doi.org/10.3109/09553002.2010.501836; PMID: 20979543
- Morgan WF. Is there a common mechanism underlying genomic instability, bystander effects and other nontargeted effects of exposure to ionizing radiation?. Oncogene 2003; 22:7094 - 9; http://dx.doi.org/10.1038/sj.onc.1206992; PMID: 14557815
- Rugo RE, Mutamba JT, Mohan KN, Yee T, Chaillet JR, Greenberger JS, et al. Methyltransferases mediate cell memory of a genotoxic insult. Oncogene 2011; 30:751 - 6; http://dx.doi.org/10.1038/onc.2010.480; PMID: 21057543
- Savage JRK. An Introduction to Chromosomal Aberrations. Atlas of Genetics and Cytogenetics in Oncology and Haematology 1999
- Kadhim MA, Lorimore SA, Townsend KM, Goodhead DT, Buckle VJ, Wright EG. Radiation-induced genomic instability: delayed cytogenetic aberrations and apoptosis in primary human bone marrow cells. Int J Radiat Biol 1995; 67:287 - 93; http://dx.doi.org/10.1080/09553009514550341; PMID: 7897277
- Zheng Q. New algorithms for Luria-Delbrück fluctuation analysis. Math Biosci 2005; 196:198 - 214; http://dx.doi.org/10.1016/j.mbs.2005.03.011; PMID: 15950991
- Dewannieux M, Esnault C, Heidmann T. LINE-mediated retrotransposition of marked Alu sequences. Nat Genet 2003; 35:41 - 8; http://dx.doi.org/10.1038/ng1223; PMID: 12897783
- Dote H, Cerna D, Burgan WE, Carter DJ, Cerra MA, Hollingshead MG, et al. Enhancement of in vitro and in vivo tumor cell radiosensitivity by the DNA methylation inhibitor zebularine. Clin Cancer Res 2005; 11:4571 - 9; http://dx.doi.org/10.1158/1078-0432.CCR-05-0050; PMID: 15958643
- Narayan A, Tuck-Muller C, Weissbecker K, Smeets D, Ehrlich M. Hypersensitivity to radiation-induced non-apoptotic and apoptotic death in cell lines from patients with the ICF chromosome instability syndrome. Mutat Res 2000; 456:1 - 15; http://dx.doi.org/10.1016/S0027-5107(00)00103-2; PMID: 11087891
- Sandhu J, Kaur B, Armstrong C, Talbot CJ, Steward WP, Farmer PB, et al. Determination of 5-methyl-2′-deoxycytidine in genomic DNA using high performance liquid chromatography-ultraviolet detection. J Chromatogr B Analyt Technol Biomed Life Sci 2009; 877:1957 - 61; http://dx.doi.org/10.1016/j.jchromb.2009.05.032; PMID: 19501556
- de Waard H, de Wit J, Gorgels TG, van den Aardweg G, Andressoo JO, Vermeij M, et al. Cell type-specific hypersensitivity to oxidative damage in CSB and XPA mice. DNA Repair (Amst) 2003; 2:13 - 25; http://dx.doi.org/10.1016/S1568-7864(02)00188-X; PMID: 12509265
- Yoder JA, Walsh CP, Bestor TH. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet 1997; 13:335 - 40; http://dx.doi.org/10.1016/S0168-9525(97)01181-5; PMID: 9260521
- Jeffreys AJ, Barber R, Bois P, Buard J, Dubrova YE, Grant G, et al. Human minisatellites, repeat DNA instability and meiotic recombination. Electrophoresis 1999; 20:1665 - 75; http://dx.doi.org/10.1002/(SICI)1522-2683(19990101)20:8<1665::AID-ELPS1665>3.0.CO;2-L; PMID: 10435430
- Klein CJ, Botuyan MV, Wu Y, Ward CJ, Nicholson GA, Hammans S, et al. Mutations in DNMT1 cause hereditary sensory neuropathy with dementia and hearing loss. Nat Genet 2011; 43:595 - 600; http://dx.doi.org/10.1038/ng.830; PMID: 21532572
- Ha K, Lee GE, Palii SS, Brown KD, Takeda Y, Liu K, et al. Rapid and transient recruitment of DNMT1 to DNA double-strand breaks is mediated by its interaction with multiple components of the DNA damage response machinery. Hum Mol Genet 2011; 20:126 - 40; http://dx.doi.org/10.1093/hmg/ddq451; PMID: 20940144
- Wang KY, James Shen CK. DNA methyltransferase Dnmt1 and mismatch repair. Oncogene 2004; 23:7898 - 902; http://dx.doi.org/10.1038/sj.onc.1208111; PMID: 15378011
- Teneng I, Montoya-Durango DE, Quertermous JL, Lacy ME, Ramos KS. Reactivation of L1 retrotransposon by benzo(a)pyrene involves complex genetic and epigenetic regulation. Epigenetics 2011; 6:355 - 67; http://dx.doi.org/10.4161/epi.6.3.14282; PMID: 21150308
- Winkelmann J, Lin L, Schormair B, Kornum BR, Faraco J, Plazzi G, et al. Mutations in DNMT1 cause autosomal dominant cerebellar ataxia, deafness and narcolepsy. Hum Mol Genet 2012; 21:2205 - 10; http://dx.doi.org/10.1093/hmg/dds035; PMID: 22328086
- Nagasawa H, Little JB. Induction of sister chromatid exchanges by extremely low doses of alpha-particles. Cancer Res 1992; 52:6394 - 6; PMID: 1423287
- Kanai Y, Ushijima S, Nakanishi Y, Sakamoto M, Hirohashi S. Mutation of the DNA methyltransferase (DNMT) 1 gene in human colorectal cancers. Cancer Lett 2003; 192:75 - 82; http://dx.doi.org/10.1016/S0304-3835(02)00689-4; PMID: 12637155
- Tufarelli C, Stanley JA, Garrick D, Sharpe JA, Ayyub H, Wood WG, et al. Transcription of antisense RNA leading to gene silencing and methylation as a novel cause of human genetic disease. Nat Genet 2003; 34:157 - 65; http://dx.doi.org/10.1038/ng1157; PMID: 12730694
- Franken NA, Rodermond HM, Stap J, Haveman J, van Bree C. Clonogenic assay of cells in vitro. Nat Protoc 2006; 1:2315 - 9; http://dx.doi.org/10.1038/nprot.2006.339; PMID: 17406473
- Carrera S, de Verdier PJ, Khan Z, Zhao B, Mahale A, Bowman KJ, et al. Protection of cells in physiological oxygen tensions against DNA damage-induced apoptosis. J Biol Chem 2010; 285:13658 - 65; http://dx.doi.org/10.1074/jbc.M109.062562; PMID: 20228054
- Duarte TL, Almeida GM, Jones GD. Investigation of the role of extracellular H2O2 and transition metal ions in the genotoxic action of ascorbic acid in cell culture models. Toxicol Lett 2007; 170:57 - 65; http://dx.doi.org/10.1016/j.toxlet.2007.02.005; PMID: 17382497
- Duarte TL, Cooke MS, Jones GD. Gene expression profiling reveals new protective roles for vitamin C in human skin cells. Free Radic Biol Med 2009; 46:78 - 87; http://dx.doi.org/10.1016/j.freeradbiomed.2008.09.028; PMID: 18973801
- Bowler DA, Moore SR, Macdonald DA, Smyth SH, Clapham P, Kadhim MA. Bystander-mediated genomic instability after high LET radiation in murine primary haemopoietic stem cells. Mutat Res 2006; 597:50 - 61; http://dx.doi.org/10.1016/j.mrfmmm.2005.04.025; PMID: 16414086
- Meng Q, Walker DM, Scott BR, Seilkop SK, Aden JK, Walker VE. Characterization of Hprt mutations in cDNA and genomic DNA of T-cell mutants from control and 1,3-butadiene-exposed male B6C3F1 mice and F344 rats. Environ Mol Mutagen 2004; 43:75 - 92; http://dx.doi.org/10.1002/em.20002; PMID: 14991748