Abstract
Mammalian oocytes contain the histone H1foo, a distinct member with low sequence similarity to other members in the H1 histone family. Oocyte-specific H1foo exists until the second embryonic cell stage. H1foo is essential for oocyte maturation in mice; however, the molecular function of this H1 subtype is unclear. To explore the function of H1foo, we generated embryonic stem (ES) cells ectopically expressing H1foo fused to an EGFP (H1foo-ES). Interestingly, ectopic expression of H1foo prevented normal differentiation into embryoid bodies (EBs). The EB preparations from H1foo-ES cells maintained the expression of pluripotent marker genes, including Nanog, Myc and Klf9, and prevented the shift of the DNA methylation profile. Because the short hairpin RNA-mediated knockdown of H1foo-EGFP recovered the differentiation ability, H1foo was involved in preventing differentiation. Furthermore, ChIP analysis revealed that H1foo-EGFP bound selectively to a set of hypomethylated genomic loci in H1foo-ES, clearly indicating that these loci were targets of H1foo. Finally, nuclease sensitivity assay suggested that H1foo made these target loci decondensed. We concluded that H1foo has an impact on the genome-wide, locus-specific epigenetic status.
Introduction
Most members of the histone H1 family bind to a stretch of linker DNA in a sequence-independent manner, connecting adjacent nucleosomes to generate higher-order chromatin structures to accurately control gene expression.Citation1,Citation2 H1foo, an oocyte-specific histone H1, is expressed during development in fully meiotically competent germinal vesicle-stage oocytes until the late two-cell embryonic stage.Citation3,Citation4 This expression is essential for oocyte maturation in miceCitation5 and has beneficial effects on developing oocytes and the fertilized egg.
H1foo shows only a very low sequence homology with somatic histone H1, though it shares a general domain structure with somatic histone H1.Citation3,Citation4 H1foo is most closely related to the oocyte-specific cleavage-stage histone H1 in sea urchin and histones B4/H1M in Xenopus laevis. Recent studies have suggested that histone B4 is involved in the nuclear reprogramming of somatic cells transferred to Xenopus oocytes via the modification of gene activities.Citation6,Citation7 However, it is still unclear whether H1foo has the same biological function as that observed in Xenopus or that observed for H1 variants that globally repress gene activity.Citation2,Citation8,Citation9
The H1foo gene is regulated by DNA methylation at tissue-dependent and differentially methylated region (T-DMR), and the expression of H1foo is stringently suppressed by DNA methylation at the T-DMR in non-expressing cells after differentiation.Citation10 Cellular differentiation alters the genome-wide epigenetic status of multiple gene loci, with changes in de novo methylation and demethylation at T-DMRs.Citation11,Citation12 The altered epigenetic status resulted in cell-type-specific DNA methylation profiles: pluripotent cells with hypomethylation of numerous T-DMRs containing genes for transcription factors (and their targets) essential for pluripotency, as well as pluripotent cells with hypermethylation of T-DMRs containing genes for tissue-specific gene expression.Citation13 The DNA methylation status of T-DMRs is closely associated with the chromatin structure, including histone modifications, and changes to histone modifications that affect the DNA methylation status and vice versa.Citation14,Citation15
We explored the link between H1foo and DNA methylation by investigating mouse embryonic stem (ES) cells with ectopic expression of H1foo on the basis of the hypothesis that H1foo influences the epigenetic status. We found that H1foo binds to chromatin and hinders the differentiation of mouse ES cells by maintaining expression of pluripotent genes through regulation of the chromatin structure.
Results
Establishment of ES cell lines ectopically expressing H1foo-EGFP
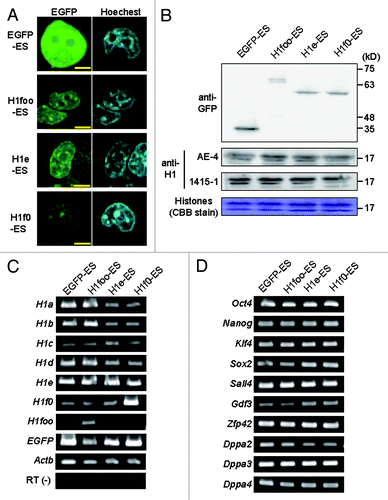
We established mouse ES cell lines stably expressing EGFP-fused mouse H1s: three cell lines expressing H1foo-EGFP (H1foo-ES) and cell lines expressing H1e-EGFP, H1f0-EGFP and EGFP (H1e-, H1f0- and EGFP-ES, respectively) as controls. In these ES cell lines, EGFP was only detected in the nucleus (not in the nucleoli) based on fluorescence images (; Fig. S1A). Western blotting of nuclear extract indicated that expression level of H1foo-EGFP was lower than those of H1e- and H1f0-EGFP (). The expression of H1s-EGFP did not affect the protein levels of endogenous H1 in nucleus, and the mRNA expression of the intrinsic H1 family genes and the ES cell marker genes (; Fig. S1B and C).
Figure 1. Establishing H1foo-EGFP expressing ES cell lines. (A) Subcellular localization of H1foo-EGFP. After wash with PBS(-), ES cells expressing H1s-EGFP fusion protein (indicated at the left side of the panels) were stained with 1 μg/ml Hoechest33342 for 10 min, and then were visualized by the confocal microscopy. The scale bars denote 5 μm. (B) The level of histone H1 proteins was evaluated by western blotting of nuclear extract prepared from EGFP-, H1foo-, H1e- and H1f0-ES cells. Endogenous and exogenous H1 were detected using anti-H1 (clone, AE-4 and 1415–1) and anti-GFP, respectively. CBB stain was used as loading control. (C) RT-PCR of H1 family genes and (D) marker genes for pluripotent stem cells in EGFP-, H1foo-, H1e- and H1f0-ES cells. RT(-) indicate PCR for Actb without reverse transcription.
H1foo-EGFP prevents differentiation
We examined the embryoid body (EB) formation in the ES cell lines to investigate the effects of H1foo on pluripotency. All of the ES cell lines formed EBs with similar sizes after culturing for 7 d. Interestingly, after culturing for 14 d, EBs derived from H1foo-ES were small and irregular-shaped compared with EBs derived from the control lines (EGFP-, H1e- and H1f0-ES) in which EBs formed yolk-sac-like structures (, Fig. S1D). These data suggested that the differentiation capacity of the H1foo-ES cell line was different from that of the control cell lines (H1e-, H1f0- and EGFP-ES).
Figure 2. H1foo-expressing ES cells exhibit defective differentiation. (A) Phase-contrast images of the EBs derived from EGFP-, H1foo-, H1e- and H1f0-ES cells after 7 and 14 d of culture. Scale bars denote 200 μm. (B) RT-PCR of maker genes for pluripotent stem cells (Zfp42, Dppa3, Nanog, Sall4 and Oct4) and markers for developmental stages (differentiation), including endoderm markers (Gata4, Gata6 and Sox17), mesoderm markers (Kdr), and an ectoderm marker (Nog), in EGFP-, H1foo-, H1e-, and H1f0-ES (U columns) and EBs cultured for 7 d (D columns). (C) RT-PCR of H1foo-EGFP-encoding transcripts during neural differentiation of H1foo-ES transfected with shRNA. Cont.-shRNA (Cont), H1foo-shRNA1(shRNA1) and H1foo-shRNA2 (shRNA2) were transfected by lipofection of H1foo-ES at 0, 3, and 6 d of culture for neural differentiation. After 10 d of culture, cells were collected, and RNA was isolated for RT-PCR. (D) Immunofluorescence images of neural differentiated H1foo-ES rescued by transfection with H1foo-shRNA. The H1foo-ES line was probed for Tubb3 (red, left), Oct4 (red, right), and DAPI (blue) after 10 d of culture. Scale bars denote 450 μm.
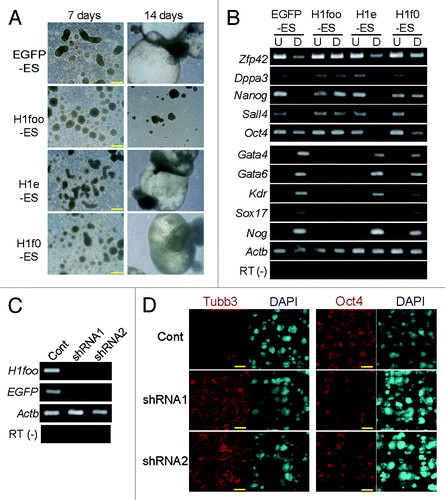
Even after 7 d, EBs derived from H1foo-ES maintained high expression levels of stem cell marker genes such as Oct4 and Nanog. In contrast, the expression of stem cell marker genes was reduced in EBs derived from the control cell lines (; Fig. S1E). In addition, differentiation marker genes, such as Gata4 and neural marker genes, were not induced in EBs and differentiated neurons derived from H1foo-ES (; Figs. S1F and 2). In contrast, overexpression of H1e and H1f0 increased expression of ectoderm and neural marker genes (Figs. S1F and 2B). These data indicated a difference between the gene regulatory network of H1foo-ES and the control cell lines.
To further confirm the role of H1foo in preventing the differentiation of H1foo-ES cells, we attempted to investigate the differentiation ability of H1foo-ES by knockdown (KD) of the H1foo-EGFP-encoding transcript using shRNA. Under culturing conditions that induced neural differentiation, the KD resulted in reduced expression of H1foo-EGFP, induced expression of the neural marker TubbIII, and reduced expression of Oct4 (). Clearly, KD of the H1foo-EGFP-encoding transcript rescued the differentiation potency to a neural lineage. H1foo-EGFP expression should thus influence the pluripotency of ES cells. We concluded that H1foo-ES cells lack pluripotency because of the ectopic expression of H1foo.
H1foo-EGFP prevents the shift of the DNA methylation profile from stem status to differentiation status
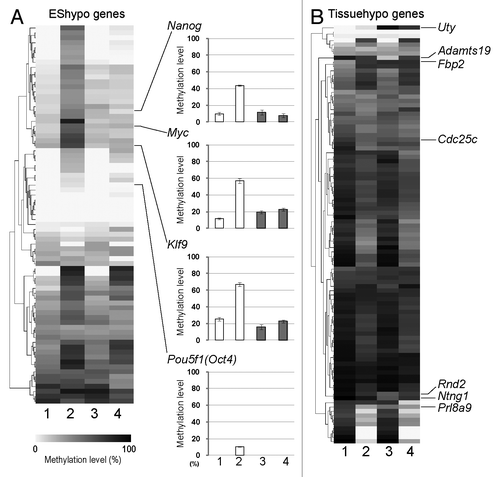
To clarify whether the DNA methylation profile is influenced by H1foo-EGFP expression, we performed combined bisulfite restriction analysis (COBRA) of EBs derived from a 7-d culture, focusing on 77 hypomethylated (EShypo T-DMRs) and 97 hypermethylated T-DMRs (Tissuehypo T-DMRs) in normal ES cells, which were selected on the basis of the previous genome-wide DNA methylation analysis of mouse ES cells.Citation13 Despite the DNA methylation profiles of H1foo-ES and EGFP-ES (; Table S1), the DNA methylation profiles of EBs derived from EGFP-ES clearly shifted from the stem state to the differentiation state (, lane 1 vs. lane 2). This change was slightly observed in EBs derived from H1foo-ES (, lane 3 vs. lane 4), suggesting that the resistance to differentiation is accompanied by the prevention of the shift of the DNA methylation profile from the stem state to the differentiation state. In contrast, similar methylation profile alterations were observed for Tissuehypo T-DMRs. Hypermethylation of 6 T-DMRs and hypomethylation of 29 T-DMRs were observed for the EBs derived from EGFP-ES and H1foo-ES (). Thus, H1foo selectively affected the DNA methylation during EB formation.
Figure 3. H1foo affects DNA methylation at specific loci. (A) DNA methylation status of EShypo (77 sites) and (B) Tissuehypo (97 sites) T-DMRs in EGFP- and H1foo-expressing ES cells and EBs cultured for 7 d. Methylation levels are visualized with a heatmap by using the MultiExperiment ViewerCitation32 and are depicted in gray-scale. For COBRA, PCR products were digested with HpyCH4IV to estimate the DNA methylation level. Data represent the mean of triplicates of cell culture. DNA methylation levels at T-DMRs in four marker genes are shown in the right panels of (A). Numbers at the bottom indicate samples: 1, EGFP-ES; 2, EB from EGFP-ES; 3, H1foo-ES; 4, EB from H1foo-ES. T-DMRs examined in this figure were selected on the basis of previous genome-wide methylation analysis.Citation13
H1foo-EGFP binds to selected EShypo T-DMR genes and Tissuehypo T-DMR genes
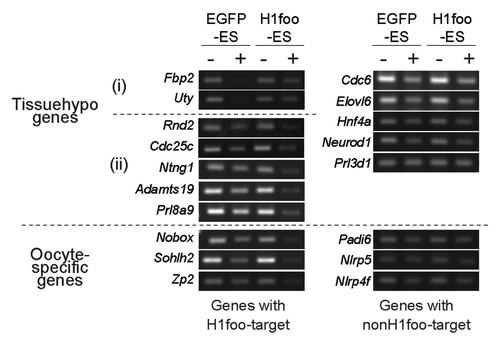
To elucidate whether H1foo binds to EShypo and Tissuehypo T-DMRs in which DNA methylation is affected by the expression of H1foo-EGFP, we performed ChIP assays using an anti-GFP antibody. T-DMRs of Nanog, Myc, and Klf9 in H1foo-ES were precipitated with anti-GFP antibodies (). We observed that the T-DMRs were resistant to DNA methylation by H1foo during the differentiation into EBs. In contrast, the Sall4- and Hsp90aa1-containing T-DMRs, where DNA methylation was not affected in H1foo-ES cells, did not precipitate. H1foo-EGFP directly bound to specific T-DMRs. We tentatively designated these T-DMRs as H1foo targets; the remaining T-DMRs were designated non-H1foo targets.
Figure 4. H1foo is localized at specific genomic loci. (A) ChIP assay of H1foo-EGFP. H1foo- and EGFP-ES cells were subjected to the ChIP assay using an antibody against GFP (P lanes). The rabbit IgG antibody was used as a negative control for the IP (C lanes) Left side of the panels indicates the type of T-DMRs. Tissuehypo genes were classified into two, indicated by (i) and (ii) according to the DNA methylation changes of T-DMRs in H1foo-ES cells. Aliquots of chromatin fragments were subjected to PCR without IP (I lanes). (B) DNA methylation status of Tissuehypo genes in EGFP-, H1foo-, H1e- and H1f0-ES cells. Bars indicate the mean values with error bars (S.E.) in triplicates of cell culture. Numbers indicate EGFP-ES,Citation1 H1foo-ES,Citation2 H1e-ESCitation3 and H1f0-ES.Citation4
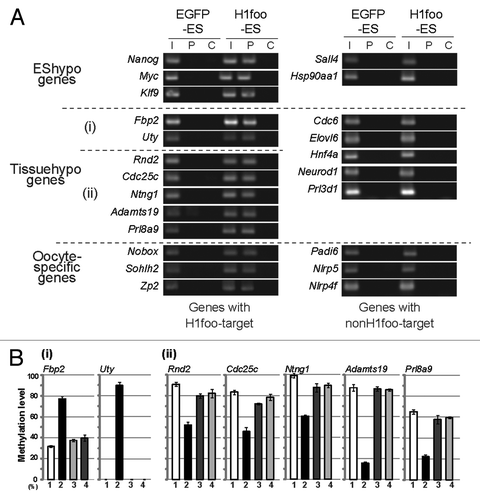
H1foo targets were found in both hypermethylated (Fbp2 and Uty) and hypomethylated (Rnd2, Cdc25c, Ntng1, Adamts19 and Prl8a9) T-DMRs in the H1foo-ES line by the ChIP assay (). H1foo targets were not detected in Tissuehypo T-DMRs (e.g., Cdc6) which DNA methylation was not affected by H1foo-EGFP expression. These ChIP data clearly indicated that H1foo-EGFP bound to specific loci to change the epigenetic status. Importantly, the DNA methylation status of these loci was not affected by the overexpression of other H1 subtypes in ES cells (). These data suggest that H1foo marks genome loci for epigenetic regulation; however, H1foo does not always contribute to the demethylation process.
H1foo-EGFP binds to selected oocyte-specific genes
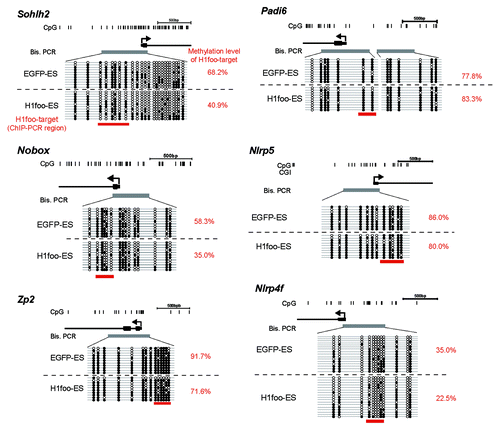
Because H1foo expression is specific to oocytes, we investigated whether H1foo-EGFP binds to T-DMRs containing certain oocyte-specific genes (Sohlh2, Nobox, Zp2, Padi6, Nlrp5 and Nlrp4f). ChIP analysis revealed that oocyte-specific genes such as Sohlh2, Nobox and Zp2 are H1foo targets (). Other oocyte-specific genes (Padi6, Nlrp5 and Nlrp4f) are not H1foo targets. We therefore speculated that there is a difference in the DNA methylation status of the H1foo targets Sohlh2, Nobox and Zp2. In these H1foo target genes, DNA was hypomethylated by more than 20% (). In contrast, the methylation status of Padi6, Nlrp5 and Nlrp4f was not affected by H1foo-EGFP expression. Increased methylation was not observed for oocyte-specific genes targeted by H1foo.
Figure 5. DNA methylation status of oocyte-specific genes in H1foo-ES cells. Top: schematic diagram of genes. The vertical lines denote the positions of cytosine residues of CG sites. The thick gray horizontal line indicates the region included in the PCR fragment. Bottom: open and filled circles represent unmethylated and methylated cytosines, respectively. The thick red horizontal lines under the circles indicate the position of the ChIP amplicon.
Nucleosome positions change with the DNA methylation status of H1foo targets
To determine the nucleosome structure of the H1foo targets, we performed a nuclease-sensitivity assay with these targets. We found that H1foo targets with induced hypomethylation (i.e., Rnd2, Cdc25c, Ntng1, Adamts19 and Prl8a9) were sensitive to nuclease treatment in H1foo-ES but not in EGFP-ES (), indicating decondensed structures. In contrast, H1foo targets with hypermethylation induction (i.e., Fbp2 and Uty), originally sensitive in EGFP-ES, were resistant to the treatment in H1foo-ES. In this experiment, non-H1foo targets were resistant to the treatment. Similarly, H1foo targets involving oocyte-specific genes (Sohlh2, Nobox and Zp2) were nuclease-sensitive only in H1foo-ES (). These data suggest that H1foo is involved in locus-specific nucleosome positioning, resulting in a DNA methylation change.
Discussion
Ectopic expression of H1foo allowed continuous expression of pluripotent marker genes such as Nanog, Myc and Klf9 under differentiation condition, and prevented differentiation of ES cells. This effect is accompanied by the inhibition of the shift in the DNA methylation profiles of multiple hypo- and hypermethylated T-DMRs. H1foo-EGFP selectively bound to a subset of T-DMRs (H1foo targets) globally distributed through the genome, thereby affecting the DNA methylation status of the ES cells. Therefore, H1foo is involved in locus-specific, global epigenetic regulation.
The involvement of histone H1 in epigenetic gene silencing and chromatin packaging has been established by monitoring the changes in the chromatin structure—because of DNA demethylation at the H19-ICR imprinting locus in the triple knockout (KO) of H1 subtypes (H1c, H1d and H1e)Citation16—as well as the strong inhibitory in vitro effect on nucleosome mobilityCitation17 and transcription.Citation18,Citation19 The chromatin architecture determines the gene activity associated with epigenetic changes.Citation14,Citation15 The chromatin structure is open at H1foo target genes; thus, H1foo is likely a biphasic epigenetic regulator.
Histone B4 is a Xenopus homolog of H1foo and is functionally different from somatic histone H1 because it influences the chromatin structure and dynamics.Citation6,Citation20 In the mouse egg, linker histones in the male pronucleus are rapidly displaced by H1foo after fertilization.Citation3 The H1 histones in the somatic nucleus are transferred to the oocytes and are displaced by H1foo.Citation21 The displacement of linker histones with B4 is also observed in mammalian somatic nuclei that express Nanog, Oct4 and Sox2, when the nuclei are transferred to Xenopus oocytes.Citation7 We showed that H1foo can maintain demethylation of T-DMRs in pluripotency-related genes as well as methylation of the T-DMRs in selected somatic genes, suggesting that H1foo is involved in epigenetic regulation in somatic nuclei and male pronuclei introduced into eggs. These data suggest the conserved molecular function between H1foo and B4.
The oocyte-specific homeobox gene Nobox regulates the expression of many genes in the ovary, including Zp2, Oct4, Gdf9, Nlrp4f and H1foo.Citation22 Another gene, Sohlh2, is required for the expression of several important oocyte-specific genes, including H1foo.Citation23 H1foo induced site-specific DNA hypomethylation in the T-DMR containing Nobox and Sohlh2. H1foo is indispensable for meiotic maturation of oocytes in mice.Citation5 These data indicated the importance of transcription factor networks and the H1foo-driven epigenetic regulation of genes for the regulation of oocyte-specific genes during oogenesis. Favorable and adverse effects that inhibit cell differentiation could define the strict expression profile of H1foo.
There are at least 7 histone H1 subtypes with different amino acid sequences in somatic cells, and the ratio of the subtypes is different in different cell types,Citation24 suggesting cell-type-dependent function and regulation. To study the role of histone H1 subtypes, however, a single gene KO was not useful for the detection of cell subtype functionCitation25 because other H1s subtypes could complement the function of the single gene. Previous studies required a triple H1-encoding gene KO in order to observe a phenotypic change.Citation20,Citation26 Despite these observations, ectopic expression of H1foo prevented cell differentiation via the blockage of a shift in the DNA methylation profile; this did not occur during normal differentiation of ES cells. In addition, the depletion of somatic H1s (H1c, H1d and H1e) prevented normal differentiation of ES cellsCitation27 and overexpression of somatic type H1s promoted differentiation of ES cells. These data indicate the import role of the displacement or replacement of H1 subtypes in normal function of cells and disorganization of H1 subtypes could be involved in pathogenesis and ectopic expression of cancer related antigens.Citation28 Thus, the expression of H1foo must be stringently regulated. In summary, H1foo prevents the shift of the DNA methylation profile following nucleosome positioning through binding to chromatin.
Materials and Methods
Plasmids
The expression vector for mouse H1foo-EGFP was kindly provided by Dr. Mamoru Tanaka.Citation21 Mouse full-length H1e- and H1f0-encoding cDNAs were isolated from total RNA from mouse ES cells by RT-PCR. The cDNA was cloned into the pGEM-T Easy vector (Promega), and the resulting constructs were confirmed by BigDye sequencing (Applied Biosystems). Cloned cDNA was inserted into pEGFP-N3 (Clontech) via the NheI and EcoRI sites to generate expression vectors for H1e- and H1f0-EGFP.
For the H1foo Knockdown (KD), a specific shRNA targeting the H1foo open reading frame was cloned into the pSingle-tTS-shRNA vector (Clontech) via the XhoI and HindIII sites. Similarly, a sequence targeting the LacZ-encoding mRNA was cloned and denoted control-sh. The shRNA sequences and the primer sequences are listed in Table S2.
Cell culture, transfection and selection of stable clones
ES cell line J1, derived from the 129S4/SvJae mouse embryo, was kindly provided by Dr. En Li.Citation29 The ES cells were cultured on a gelatin-coated dish (Sigma-Aldrich) in the presence of 1500 U/ml leukemia inhibitory factor (ESGRO; Millipore) under standard conditions.Citation30 Cells were cultured in 6-well dishes to 50% confluence and were then transfected with 5 μg of plasmid and 5 μl of Lipofectamine 2000 (Invitrogen) per dish. Twenty-four hours after transfection, cells were replated and cultured for a week in the presence of 300 μg/ml G418 (Invitrogen) in a 10-cm dish. G418-resistant colonies were transferred to a 96-well plate. Cells that expressed the fusion proteins were collected, frozen in liquid nitrogen, and stored in Cell Culture Freezing Media (Cell Banker1; JUJI FIELD INC.) at -80°C until use.
Embryoid body formation and neural differentiation of ES cells
For EB formation, 3 × 106 cells were cultured in 10-cm bacteriological dishes in the absence of ESGRO, and the medium was exchange every 2 d. For neural differentiation, PA6 cells were plated onto a 10-cm dish and cultured for 5 d in Minimum Essential Medium (MEM) α (Invitrogen) supplemented with 10% FBS. After the cells were confluent, the cell surface was washed three times with 10 ml of PBS(-) to minimize the contamination of FBS. Subsequently, ES cells (1 × 105) were cultured on PA6 feeder cells in Glasgow MEM (Invitrogen) supplemented with 10% Knockout Serum Replacement Medium (Invitrogen), 0.1 mM nonessential amino acids (Invitrogen), and 0.1 mM β-mercaptoethanol (Invitrogen). The culture medium was exchanged on day 4 and every 2 d thereafter.
RT-PCR
Total RNA was isolated from cells and tissues with the TRIzol reagent (Invitrogen) according to the manufacturer’s instructions. For RT-PCR analysis, first-strand cDNA was synthesized from 3 μg of total RNA by using the oligo(dT)20 primer and the SuperScript III First-Strand Synthesis System for RT-PCR (Invitrogen). The PCR was conducted with LA Taq DNA polymerase (Takara). PCR reactions were performed under the following conditions: denaturation at 95°C for 3 min and 30 cycles (only Actb, 20 cycles), each cycle comprising 95°C for 30 sec, 60°C for 30 sec, 72°C for 15 sec. PCR products were subjected to agarose gel electrophoresis and stained using ethidium bromide.
RT-qPCR
We performed RT-qPCR using a high throughput gene expression platform based on microfluidic dynamic arrays (Fluidigm). A single aliquot of each cDNA sample, equivalent to 12.5 ng RNA, was pre-amplified using TaqMan PreAmp Mastermix (Applied Biosystems) according to manufacturer's protocol. Following pre-amplification, the samples were diluted 1:5 in TE buffer (pH 8.0). Primers and Universal probes were purchased from Roche (Table S2). BioMark 48x48 arrays were prepared according to the manufacturer's instructions. Following loading of the assays and samples into the chip by the IFC controller, PCR was performed with the following reactions conditions: 50°C for 2 min, 95°C for 10 min, followed by 40 cycles of 95°C for 15 sec and 60°C for 60 sec. Data was processed by automatic global threshold setting with the same threshold value for all assays and linear baseline correction using BioMark Real-time PCR Analysis software.
Immunocytochemical analysis
Cells cultured in 4-well dishes were fixed with 4% paraformaldehyde (Wako) and permeabilized with 0.2% Triton X-100 (Wako) followed by blocking with 5% BSA (Sigma) and incubation with the primary antibody overnight at 4°C. The secondary antibody was added and the incubation was continued for 1 h at room temperature. Nuclei were stained with DAPI (1 μg/ml; Wako). The following primary antibodies were used: mouse anti-Tubb3 (1:500; Covance), and mouse anti-Oct4 (1:200; Santa Cruz). The following secondary antibodies were used goat anti-mouse Alexa-Flour 594 (1:1000; Invitrogen). Fluorescence images were acquired with a confocal microscope (CellVoyager CV1000; Yokogawa).
Western blotting
Nuclear fractions of each sample were collected using Nuclear Extract Kit (Active Motif) according to the manufacturer’s protocols. The proteins were fractionated by 5–20% SDS-PAGE (XV PANTERA Gel; DRC), blotted onto nitrocellulose membranes and incubated at 4°C with mouse anti-GFP (B-2, 1:200; Santa Cruz), mouse anti-H1 (AE-4, 1:500; Santa Cruz) and mouse anti-H1 Carboxyterminal end (1415–1, 1:200; Abcam) antibodies. Protein bands were detected using secondary antibody coupled to horseradish peroxidase (Jackson ImmunoResearch) and then SuperSignal West Pico (Thermo).
DNA methylation analysis using the bisulfite method
Genomic DNA was extracted from ES cells as described previously.Citation31 Genomic DNA was digested with HindIII (Takara), and 5 μg of digested DNA was denatured with 0.3 M NaOH. Sodium metabisulfite (pH 5.0) and hydroquinone were added at final concentrations of 2.0 M and 0.5 mM, respectively. The reaction mixture was incubated under the following conditions: 15 cycles of 95°C for 30 sec and 50°C for 15 min. Next, DNA was purified using the gel extraction kit (QIAGEN), eluting in 100 μl of elution buffer. DNA was treated with 0.3 M NaOH at 37°C for 15 min, precipitated with 6 M ammonium acetate (pH 7.0) and ethanol, and dissolved in 200 μl of TE (pH 8.0). For each bisulfite PCR, 2 μl of DNA solution was used as a template, and BIOTAQ HS DNA polymerase (Bioline) was used to catalyze the amplification. PCR was performed with the following thermocycling conditions: denaturation at 95°C for 10 min and 43 cycles, each cycle comprising incubation at 95°C for 30 sec, 60°C for 45 sec, and 72°C for 30 sec, followed by a final extension for 10 min at 72°C.
For COBRA, PCR products were digested with HpyCH4IV (New England Biolabs). Restriction enzyme-treated DNA was purified by gel filtration using Sephadex G-5 (GE Healthcare), and the concentrations (ng/ml) of cut (derived from methylated DNA) and uncut fragments (derived from unmethylated DNA) were quantified by MultiNA Microchip Electrophoresis (Shimadzu). The methylation level was calculated as the concentration ratio of the cut fragments to the cut plus uncut fragments. For sequencing, the PCR fragments were cloned into the pGEM-T Easy vector (Promega).
ChIP assay
The ChIP assay was performed with 1 × 107 cells per assay by using the ChIP-IT Express Kit (Active Motif) according to the manufacturer’s instructions. Briefly, fixed cells were lysed and mixed with an enzymatic shearing cocktail for 10 min. Anti-GFP antibodies (Abcam) were used for IP, and rabbit IgG antibodies (Abcam) were used as a negative control to determine the IP specificity. After IP, recovered DNA was subjected to PCR with LA Taq DNA polymerase. PCR reactions were performed under the following conditions; denaturation at 95°C for 3 min and 30 cycles, each cycle comprising 95°C for 30 sec, 60°C for 30 sec, 72°C for 15 sec. PCR products were subjected to agarose gel electrophoresis and stained using ethidium bromide.
Nuclease-sensitivity assay
The nuclease-sensitivity assay can be used to determine the sensitivity of DNA regions to nucleases. This assay can thus reveal the location of open or closed chromatin. The method is based on the more rapid nuclease-catalyzed hydrolysis observed with open chromatin structures than with closed chromatin structures. Nuclease-sensitivity assays were performed with 2 × 106 cells per assay by using the EpiQ Chromatin Analysis Kit (Bio-Rad) according to the manufacturer’s instructions. Briefly, living cells were lysed and sheared with the EpiQ enzyme for 1 h. DNA was then purified and used for PCR with LA Taq DNA polymerase. PCR reactions were performed under the same conditions as ChIP assay.
| Abbreviations: | ||
| EGFP | = | enhanced green fluorescent protein |
| PBS | = | phosphate buffered saline |
| BSA | = | bovine serum albumin |
| CBB | = | Coomassie Brilliant Blue |
| DAPI | = | 4',6-diamidino-2-phenylindole |
| RT | = | reverse transcription |
| qPCR | = | quantitative polymerase chain reaction |
| COBRA | = | combined bisulfite restriction analysis |
| ChIP | = | chromatin immune precipitation |
| ES | = | embryonic stem |
| T-DMR | = | tissue-dependent and differentially methylated region |
Additional material
Download Zip (981 KB)Acknowledgments
This research was funded by a Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology (MEXT), Japan (no. 21221008 to K.S. and no.10014250 to J.O.), as well as a grant from the National Institute of Biomedical Innovation (NIBIO), Japan. The authors declare no conflict of interest. We acknowledge Dr. Yoshikazu Arai, Mr. Masaki Takasugi, Ms. Momo O. Nakanishi and Mr. Yasuyuki Tabei for technical assistance. The author contributions are as follows: Koji Hayakawa, Shintaro Yagi and Kunio Shiota designed this study. The study was discussed with Jun Ohgane and Satoshi Tanaka. Koji Hayakawa, Jun Ohgane and Satoshi Tanaka generated the H1s-expressing ES cell lines. Koji Hayakawa and Jun Ohgane performed the bisulfite experiments. Koji Hayakawa performed all other experiments. Koji Hayakawa, Shintaro Yagi and Kunio Shiota prepared the manuscript.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental Material
Supplemental materials may be found here: www.landesbioscience.com/journals/epigenetics/article/21492
References
- Hansen JC. Conformational dynamics of the chromatin fiber in solution: determinants, mechanisms, and functions. Annu Rev Biophys Biomol Struct 2002; 31:361 - 92; http://dx.doi.org/10.1146/annurev.biophys.31.101101.140858; PMID: 11988475
- Bustin M, Catez F, Lim JH. The dynamics of histone H1 function in chromatin. Mol Cell 2005; 17:617 - 20; http://dx.doi.org/10.1016/j.molcel.2005.02.019; PMID: 15749012
- Tanaka M, Hennebold JD, Macfarlane J, Adashi EY. A mammalian oocyte-specific linker histone gene H1oo: homology with the genes for the oocyte-specific cleavage stage histone (cs-H1) of sea urchin and the B4/H1M histone of the frog. Development 2001; 128:655 - 64; PMID: 11171391
- Tanaka M, Kihara M, Meczekalski B, King GJ, Adashi EY. H1oo: a pre-embryonic H1 linker histone in search of a function. Mol Cell Endocrinol 2003; 202:5 - 9; http://dx.doi.org/10.1016/S0303-7207(03)00054-6; PMID: 12770723
- Furuya M, Tanaka M, Teranishi T, Matsumoto K, Hosoi Y, Saeki K, et al. H1foo is indispensable for meiotic maturation of the mouse oocyte. J Reprod Dev 2007; 53:895 - 902; http://dx.doi.org/10.1262/jrd.19008; PMID: 17519519
- Saeki H, Ohsumi K, Aihara H, Ito T, Hirose S, Ura K, et al. Linker histone variants control chromatin dynamics during early embryogenesis. Proc Natl Acad Sci U S A 2005; 102:5697 - 702; http://dx.doi.org/10.1073/pnas.0409824102; PMID: 15821029
- Jullien J, Astrand C, Halley-Stott RP, Garrett N, Gurdon JB. Characterization of somatic cell nuclear reprogramming by oocytes in which a linker histone is required for pluripotency gene reactivation. Proc Natl Acad Sci U S A 2010; 107:5483 - 8; http://dx.doi.org/10.1073/pnas.1000599107; PMID: 20212135
- Sera T, Wolffe AP. Role of histone H1 as an architectural determinant of chromatin structure and as a specific repressor of transcription on Xenopus oocyte 5S rRNA genes. Mol Cell Biol 1998; 18:3668 - 80; PMID: 9632749
- Lee H, Habas R, Abate-Shen C. MSX1 cooperates with histone H1b for inhibition of transcription and myogenesis. Science 2004; 304:1675 - 8; http://dx.doi.org/10.1126/science.1098096; PMID: 15192231
- Maeda C, Sato S, Hattori N, Tanaka S, Yagi S, Shiota K. DNA hypomethylation circuit of the mouse oocyte-specific histone H1foo gene in female germ cell lineage. Biol Reprod 2008; 78:816 - 21; http://dx.doi.org/10.1095/biolreprod.107.066522; PMID: 18184919
- Kremenskoy M, Kremenska Y, Ohgane J, Hattori N, Tanaka S, Hashizume K, et al. Genome-wide analysis of DNA methylation status of CpG islands in embryoid bodies, teratomas, and fetuses. Biochem Biophys Res Commun 2003; 311:884 - 90; http://dx.doi.org/10.1016/j.bbrc.2003.10.078; PMID: 14623263
- Sakamoto H, Suzuki M, Abe T, Hosoyama T, Himeno E, Tanaka S, et al. Cell type-specific methylation profiles occurring disproportionately in CpG-less regions that delineate developmental similarity. Genes Cells 2007; 12:1123 - 32; http://dx.doi.org/10.1111/j.1365-2443.2007.01120.x; PMID: 17903172
- Sato S, Yagi S, Arai Y, Hirabayashi K, Hattori N, Iwatani M, et al. Genome-wide DNA methylation profile of tissue-dependent and differentially methylated regions (T-DMRs) residing in mouse pluripotent stem cells. Genes Cells 2010; 15:607 - 18; http://dx.doi.org/10.1111/j.1365-2443.2010.01404.x; PMID: 20477876
- Li E. Chromatin modification and epigenetic reprogramming in mammalian development. Nat Rev Genet 2002; 3:662 - 73; http://dx.doi.org/10.1038/nrg887; PMID: 12209141
- Ikegami K, Ohgane J, Tanaka S, Yagi S, Shiota K. Interplay between DNA methylation, histone modification and chromatin remodeling in stem cells and during development. Int J Dev Biol 2009; 53:203 - 14; http://dx.doi.org/10.1387/ijdb.082741ki; PMID: 19412882
- Fan Y, Nikitina T, Zhao J, Fleury TJ, Bhattacharyya R, Bouhassira EE, et al. Histone H1 depletion in mammals alters global chromatin structure but causes specific changes in gene regulation. Cell 2005; 123:1199 - 212; http://dx.doi.org/10.1016/j.cell.2005.10.028; PMID: 16377562
- Pennings S, Meersseman G, Bradbury EM. Linker histones H1 and H5 prevent the mobility of positioned nucleosomes. Proc Natl Acad Sci U S A 1994; 91:10275 - 9; http://dx.doi.org/10.1073/pnas.91.22.10275; PMID: 7937940
- Laybourn PJ, Kadonaga JT. Role of nucleosomal cores and histone H1 in regulation of transcription by RNA polymerase II. Science 1991; 254:238 - 45; http://dx.doi.org/10.1126/science.1718039; PMID: 1718039
- Shimamura A, Sapp M, Rodriguez-Campos A, Worcel A. Histone H1 represses transcription from minichromosomes assembled in vitro. Mol Cell Biol 1989; 9:5573 - 84; PMID: 2586527
- Godde JS, Ura K. Dynamic alterations of linker histone variants during development. Int J Dev Biol 2009; 53:215 - 24; http://dx.doi.org/10.1387/ijdb.082644jg; PMID: 19247968
- Rajkovic A, Pangas SA, Ballow D, Suzumori N, Matzuk MM. NOBOX deficiency disrupts early folliculogenesis and oocyte-specific gene expression. Science 2004; 305:1157 - 9; http://dx.doi.org/10.1126/science.1099755; PMID: 15326356
- Choi Y, Yuan D, Rajkovic A. Germ cell-specific transcriptional regulator sohlh2 is essential for early mouse folliculogenesis and oocyte-specific gene expression. Biol Reprod 2008; 79:1176 - 82; http://dx.doi.org/10.1095/biolreprod.108.071217; PMID: 18753606
- Zhang Y, Cooke M, Panjwani S, Cao K, Krauth B, Ho PY, et al. Histone h1 depletion impairs embryonic stem cell differentiation. PLoS Genet 2012; 8:e1002691; http://dx.doi.org/10.1371/journal.pgen.1002691; PMID: 22589736
- Wang J, Emadali A, Le Bescont A, Callanan M, Rousseaux S, Khochbin S. Induced malignant genome reprogramming in somatic cells by testis-specific factors. Biochim Biophys Acta 2011; 1809:221 - 5; http://dx.doi.org/10.1016/j.bbagrm.2011.04.003; PMID: 21530697
- Khochbin S. Histone H1 diversity: bridging regulatory signals to linker histone function. Gene 2001; 271:1 - 12; http://dx.doi.org/10.1016/S0378-1119(01)00495-4; PMID: 11410360
- Zlatanova J, Caiafa P, Van Holde K. Linker histone binding and displacement: versatile mechanism for transcriptional regulation. FASEB J 2000; 14:1697 - 704; http://dx.doi.org/10.1096/fj.99-0869rev; PMID: 10973918
- Fan Y, Nikitina T, Morin-Kensicki EM, Zhao J, Magnuson TR, Woodcock CL, et al. H1 linker histones are essential for mouse development and affect nucleosome spacing in vivo. Mol Cell Biol 2003; 23:4559 - 72; http://dx.doi.org/10.1128/MCB.23.13.4559-4572.2003; PMID: 12808097
- Teranishi T, Tanaka M, Kimoto S, Ono Y, Miyakoshi K, Kono T, et al. Rapid replacement of somatic linker histones with the oocyte-specific linker histone H1foo in nuclear transfer. Dev Biol 2004; 266:76 - 86; http://dx.doi.org/10.1016/j.ydbio.2003.10.004; PMID: 14729479
- Li E, Bestor TH, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell 1992; 69:915 - 26; http://dx.doi.org/10.1016/0092-8674(92)90611-F; PMID: 1606615
- Matise MP, Auerbach W, Joyner AL. (2000) in Gene Targeting: A Practical Approach, ed Joyner AL (Oxford University Press), pp 102-132.
- Yagi S, Hirabayashi K, Sato S, Li W, Takahashi Y, Hirakawa T, et al. DNA methylation profile of tissue-dependent and differentially methylated regions (T-DMRs) in mouse promoter regions demonstrating tissue-specific gene expression. Genome Res 2008; 18:1969 - 78; http://dx.doi.org/10.1101/gr.074070.107; PMID: 18971312
- Chu VT, Gottardo R, Raftery AE, Bumgarner RE, Yeung KY. MeV+R: using MeV as a graphical user interface for Bioconductor applications in microarray analysis. Genome Biol 2008; 9:R118; http://dx.doi.org/10.1186/gb-2008-9-7-r118; PMID: 18652698