Abstract
Sperm chromatin reveals two characteristic features in that protamines are the predominant nuclear proteins and remaining histones are highly acetylated. Histone H4 acetylated at lysine 12 (H4K12ac) is localized in the post-acrosomal region, while protamine-1 is present within the whole nucleus. Chromatin immunoprecipitation in combination with promoter array analysis allowed genome-wide identification of H4K12ac binding sites. Previously, we reported enrichment of H4K12ac at CTCF binding sites and promoters of genes involved in developmental processes. Here, we demonstrate that H4K12ac is enriched predominantly between ± 2 kb from the transcription start site. In addition, we identified developmentally relevant H4K12ac-associated promoters with high expression levels of their transcripts stored in mature sperm. The highest expressed mRNA codes for testis-specific PHD finger protein-7 (PHF7), suggesting an activating role of H4K12ac in the regulatory elements of this gene. H4K12ac-associated genes revealed a weak correlation with genes expressed at 4-cell stage human embryos, while 23 H4K12ac-associated genes were activated in 8-cell embryo and 39 in the blastocyst. Genes activated in 4-cell embryos are involved in gene expression, histone fold and DNA-dependent transcription, while genes expressed in the blastocyst were classified as involved in developmental processes. Immunofluorescence staining detected H4K12ac from the murine male pronucleus to early stages of embryogenesis. Aberrant histone acetylation within developmentally important gene promoters in infertile men may reflect insufficient sperm chromatin compaction, which may result in inappropriate transfer of epigenetic information to the oocyte.
Introduction
At fertilization, human sperm chromatin carries both genetic and epigenetic information that is essential for proper embryo development. Our current state of knowledge points to different structural domains of sperm chromatin, each relating to a different function. The largest structural domain is made up of arginine-rich, protamine-bound sperm DNA, which replaces > 85% of somatic histones and renders the paternal genome transcriptionally inactive. In addition, formation of stacked, protamine-DNA toroids increases the compaction of the paternal genome. The stabilization of this tightly packaged chromatin occurs through intermolecular disulfide cross-links. A newly proposed model of toroid stability includes the formation of zinc bridges with protamine thiols of cysteine and potential imidazole groups of histidine.Citation1 Another sperm chromatin domain contains approximately 15% of the remaining histones that are comprised of canonical histones (H2A, H2B, H3, H4) as well as a testis-specific histone variant (tH2B).Citation2,Citation3 Histone tails contain residues that can be post-translationally modified by methylation, acetylation, phosphorylation, ubiquitylation and sumoylation.Citation4 These post-translational modifications alter chromatin structure, thereby allowing the underlying gene to be activated or repressed. Retained sperm nucleosomes are either passive remnants of incomplete histone to protamine replacement, or may play an active and biologically relevant role during early embryogenesis.
Shortly after fertilization, the protamines in sperm chromatin are replaced with histones supplied by the oocyte,Citation5-Citation7 but in those regions where histones are already present in the sperm DNA, this may not be necessary. Van der Heijden et al.Citation6,Citation8 demonstrated that histones with specific modifications in the sperm cell are also present in the paternal pronucleus suggesting that they originate in the sperm. The transmission of sperm histones, and the associated chromatin structures, suggest that it is possible that the newly fertilized oocytes inherit histone-based chromatin structural organization from the sperm.
Van der Heijden et al.Citation6 observed histone H4 acetylated at K8 and K12 prior to full decondensation of the sperm nucleus suggesting that these marks are transmitted by the spermatozoon. Tracking down the origin of H4K8ac and H4K12ac during spermiogenesis revealed the retention of nucleosomes with these modifications in the chromocenter of elongating spermatids. The authors showed that constitutive sperm heterochromatin is enriched for nucleosomes carrying specific histone modifications, which are transmitted to the zygote. In general, acetylated histone residues are involved in the activation of gene expression. Immunofluorescence does not provide information on the identity of DNA binding sites associated with these epigenetic marks in the paternal genome. However, as such activating modifications binding to regulatory gene elements are located close to the transcription start sites (TSS), it is possible that activating epigenetic marks on paternal chromatin (acetylated histone) will initiate the transcription machinery after fertilization.
In a previous study,Citation9 we reported a comparison between sperm H4K12ac chromatin immunoprecipitation (ChIP) and the chromatin fraction isolated using high salt and endonuclease digestion of sperm chromatin. We demonstrated that the DNA sequence composition of soluble chromatin was qualitatively similar to H4K12ac ChIP chromatin and included a strong developmental ontology. In the present study, we applied ChIP followed by promoter microarray analysis to identify H4K12ac-associated gene promoters in sperm DNA from healthy donors. We analyzed averaged data from ejaculates in terms of distance of binding to the transcription start sites and compared H4K12ac interacting genes with mRNA profiles in sperm and gene expression profiling from human pre-implantation embryos.
Results
Identification of fertility-related and developmentally important gene promoters interrogating with H4K12ac
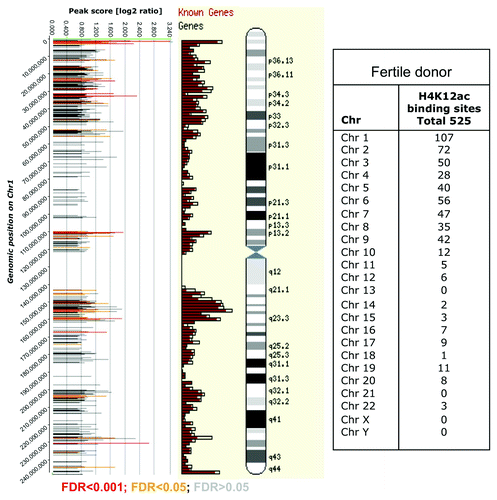
Using CHIP-on-chip we have identified 514 gene promoters as potential binding sites for anti-H4K12ac antibody. The promoter for PEX26 on chromosome 22 was identified as the highest peak (with a maximum level of 3.3 after averaging and normalizing our data; ; Fig. S1). A higher binding frequency was detected on telomeric fragments of large chromosomes (chr 1 – chr 9). The distribution of binding sites in sperm of one fertile donor is shown in .
Figure 1. Assessment of microarray data quality by ArrayQualityMetrics package of Bioconductor and normalization of biological replicates. A. Scatterplot and Spearmann correlation of raw intensities between two biological replicates. The ArrayQualityMetrics tool allowed generation of correlation co-efficiency value between ChIP-on-chip replicates CC = 0.783 (0, no correlation; 1, perfect correlation). (B) Histogram of mean signal ratios of H4K12ac chromatin-immunoprecipitated DNA vs. random-sheared total genomic DNA (Input). The distinct tail at the right-hand end corresponds to DNA fragments enriched by H4K12ac. Tiled oligos that displayed the top 3% ratios are located to the right of the red bar. (C) The highest peak obtained after averaging of 2 replicates as calculated by log ratios between input material and immunoprecipitated sample.
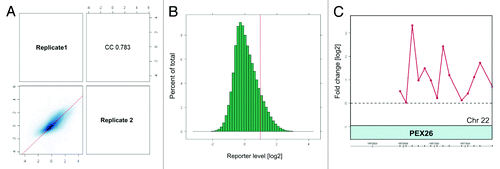
Figure 2. Distribution of H4K12ac peaks (shown as bars) along chromosome 1 from one healthy donor aligned alongside Ensembl gene density profile and ideograms of chromosome 1. The positions of the H4K12ac peaks on were plotted against their enrichment, which was calculated from the log2-ratio of input signal-Cy3 for the total chromatin (control) and IP-probe–Cy5. False discovery rate (FDR) predicts significance of peak enrichment. (Red bars; p < 0.001, yellow bars; p < 0.05, gray bars p > 0.05). Higher density of binding sites was recognized in telomeric sequences and region of high gene density on chromosome 1. Table summarizes the frequency of binding of all human chromosomes.
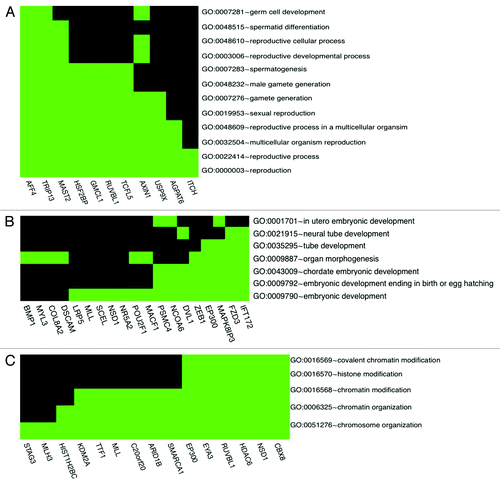
Gene ontology classification has enabled us to highlight specific anti-H4K12ac-bound promoters that may be sperm specific and may also be important later during embryonic development. Eleven genes were classified to be involved in reproductive processes (GO:0022414) (): AGPAT6, AFF4, RUVBL1, AXIN1, GMCL1, HSF2BP, ITCH, MAST2, TRIP13, TCFL5 and USP9X. Among those genes AF4/FMR2 family, member 4 (AFF4), microtubule associated serine/threonine kinase 2 (MAST 2) and thyroid hormone receptor interactor 13 (TRIP13) play roles in spermatid differentiation and may be important for male fertility. For instance, Urano et al.Citation10 reported that AFF4 (AF5q31)-deficient male mice displayed spermatogenic arrest and developed azoospermia. Furthermore, impairment of protamine-1, protamine-2 and transition protein-2 expression have been observed in AFF4 mutant mice. This finding raises the possibility that AFF4 also directly regulates the transcription of protamine genes, which are required to condense the haploid genome within the sperm nucleus. It could also be the case that aberrant histone modification of the AFF4 promoter, more precisely, H4K12 deacetylation during spermatogenesis, may also be related to male infertility. Interestingly, we found 15 anti-H4K12ac-bound gene promoters relating to embryonic development (GO:0009790), of which 8 (EP300, DVL1, FZD3, IFT172, MAPK8IP3, NCOA6, PSMC4, ZEB1) () were classified according to embryonic development ending in birth or in egg hatching (GO:0009792); for example, EP300 (E1A binding protein p300) is a transcriptional co-activator with intrinsic histone acetylase activity, which contributes to transcriptional activation.Citation11 There is evidence that Pcaf (p300) is dispensable for murine development.Citation12 In contrast, Gcn5l2-null embryos develop normally up to 7.5 d post coitum (d.p.c.), but their growth is severely retarded by 8.5 d.p.c. and they fail to form dorsal mesoderm lineages, including chordamesoderm and paraxial mesoderm. The interaction of H4K12ac with p300 acetyltransferase may predict proper embryo development, but on the other hand, lack of p300 promoter occupancy by H4K12ac in sperm may lead to embryo miscarriage and abortion. The following gene promoters have also been classified to chromatin modification process (GO:0006325), which may predict their regulatory role after fertilization: STAG3, MLH3, HIST1H2BC, KDM2A, HDAC6, NSD1, CBX8 ().
Figure 3. H4K12ac-associated promoters involved in reproductive process (A), embryonic development (B) and chromatin organization (C). Spermatogenesis / fertility-related and developmentally important gene promoters selected according to gene ontology classification available as a web tool (http://david.abcc.ncifcrf.gov/).
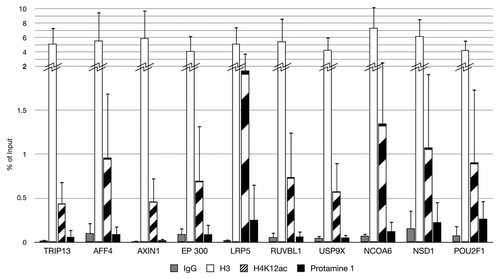
Validation of CHIP-on-chip enriched regions
We chose 10 fertility and developmentally important promoter candidates with significantly high enrichment scores for evaluation of the H4K12ac microarray data. Identified peaks within the gene promoters TRIP13, AFF4, AXIN1, EP300, LRP5, RUVBL1, USP9X, NCOA6, NSD1 and POU2F1 are shown in . Our result showed high (2–5-fold) enrichment for H4K12ac for investigated binding sites within selected promoters in ejaculates from three different healthy donors. However, strong individual differences could be observed (high standard deviation). The enrichment of H4K12ac was 4-fold higher than protamine 1 in each donor. This result confirmed for LPR5 as a binding site for anti-H4K12ac in sperm of healthy men. However, when compared with a group of subfertile patients, the enrichment on LPR5 was strongly reduced (0.2–0.5-fold). This reduction was compensated by unmodified H3, with strong individual differences being detected (data not shown).
Distance of H4K12ac enrichment sites to the transcription start site (TSS)
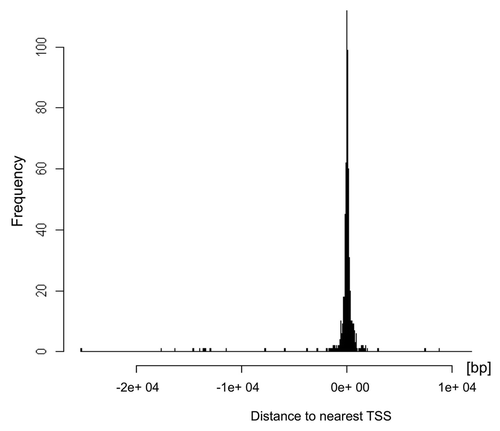
In somatic cells, induced genes tend to be bound by active marks like transcription factors and acetylated histones to the regulatory elements near to transcription start site. However, in spermatozoa, transcription is terminated through a high compaction of the chromatin and protamine interaction. Thus, we assume that the distance of binding of anti-H4K12ac to TSS of certain genes in sperm may predict their transcriptional potential after fertilization. We therefore examined more thoroughly the pattern of H4K12ac occupancy in the region of 9 kb upstream and 5 kb downstream from the TSS. illustrates the frequency of binding for anti-H4K12ac near the transcription start sites. Our results indicate that the enriched intensities in relation to the gene start coordinate are, on average, highest between 0–2 kb upstream from the TSS. Providing that the sperm genome is marked with H4K12ac in the region spanning from 2 kb upstream to 2 kb downstream of the TSS, it is more likely that the expression of those genes might be induced after fertilization.
Figure 5. Distribution of H4K12ac binding sites relative to transcription start sites (TSS) of the spermatozoal genome. Histogram generated using ChIPpeakAnno package of R software shows that H4K12ac interacts with regions symmetrically distributed around TSS. The highest binding frequency could be observed 2 kb upstream and 2 kb downstream from TSS.
Co-localization of H4K12ac with H3K9ac, H3K4me3 and H3K27me3 in human spermatozoa
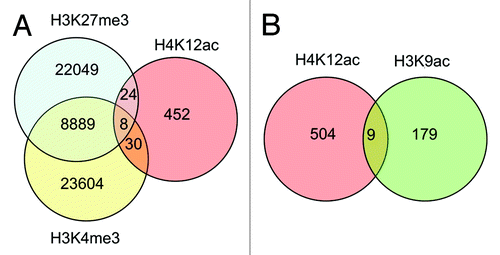
Comparing data from promoter array with anti-H4K12ac with data of anti-H3K9ac CHIP-on–chip ENCODE array in fertile donors,Citation13 co-localization of binding sites for anti-H4K12ac and anti-H3K9ac could be demonstrated for 9 gene promoters that are involved in reproductive processes and embryonic development, e.g., AFF4 (AF4/FMR2 family, member 4), AXIN1, NCOA6 (nuclear receptor co-activator 6), OR52A1 (olfactory receptor, family 52, subfamily A, member 1). To compare our H4K12ac data set with H3K27me3 and H3K4me3 we used pre-compiled peak list from Hammoud et al.Citation14 Peaks with promoter occupancy have been extracted from Chip-seq reads. We found out very minor overlap between those histone modifications. Our results showed that bivalent interaction of H4K12ac and H3K4me3 has been observed in 30 gene promoters. H3K27me3 and H4K12ac have been found in 24 promoters. All three modifications were found in 8 promoters. Considering an enormous data set for H3K27me and H3K4me3, H4K12ac represent a very low fraction (;Fig. S2).
Figure 6. Venn diagram demonstrating co-localization of H4K12ac with methylated histones: H3K27me3, and H3K4me3 (A), and H3K9ac (B) within promoters of human sperm. Analysis was performed with data published by Hammoud et al.Citation14 and ENCODE CHIP-on Chip with H3K9ac from our previous study.Citation13
Relationship between H4K12ac promoter occupancy and sperm mRNA level
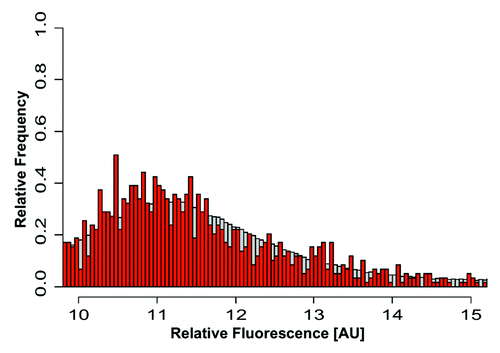
H4K12ac is an activating epigenetic mark, which, alongside the mRNAs stored in mature spermatozoa, may represent a useful fingerprint for monitoring past events, especially the developmental profile of gene expression during spermatogenesis. In order to investigate whether promoter association with H4K12ac in specific promoters is possibly correlated with mRNA transcripts stored in spermatozoa, we compared data from ChIP-on-chip of H4K12ac with genome wide expression profiling of mRNAs from human sperm (CodeLink Human Whole Genome Bioarray, Applied Microarrays). Among the 514 gene promoters that have been identified as binding sites for H4K12ac, we found 98 mRNA transcripts identified by genome-wide (relative expression level). Interestingly, H4K12ac interacting genes display high level of mRNA transcripts in spermatozoa. Our analysis showed that 94 H4K12ac associated gene promoters have mRNA expression levels between 10–14 relative fluorescence units (; Fig. S2). There was no correlation between peak score for H4K12ac (probability of binding site) and relative fluorescence corresponding to mRNA level (p = 0.2873).
Figure 7. mRNA expression levels of H4K12ac-associated genes in spermatozoa. The plot demonstrates the distribution (as relative frequencies, ordinate) of microarray derived relative mRNA expression levels (abscissa) in the group of H4K12ac-associated genes (red bars) compared with the group of all 20,400 genes (gray bars) from fertile donors. The list of H4K12ac-associated promoters with corresponding mRNA transcripts in sperm of healthy donors are shown in Figure S2.
The highest expressed mRNAs are coding testis-specific proteins, such as PHF7 (PHD finger protein 7, Testis development protein NYD-SP6; mRNA level 15) and EHD1 (EH domain-containing protein 1, Testilin, (hPAST1); mRNA level 14). Differentially expressed genes with promoters enriched in H4K12ac in sperm chromatin were subjected to gene ontology classification and clustering with the DAVID2010 database. Functional terms such as “cellular protein molecular process” (35 out of 98; p value = 3.1e-4), “positive regulation of transcription, DNA-dependent” (11 out of 98; p value = 5.4e-3), “embryonic development” (11 out of 98; p value = 6.4e-2) are overrepresented in the group of genes, possibly expressed through acetylation of H4K12 during spermatogenesis.
Expression of sperm H4K12ac-marked genes during early embryo development
We compared transcriptome data from pre-implantation embryosCitation12 with ChIP-on-chip of H4K12ac from sperm in order to analyze whether H4K12ac-marked genes in sperm possibly enhance their transcriptional activity after fertilization. This analysis provides additional information about which maturation phase in which H4K12ac-marked genes are induced to be transcribed. Zhang et al.Citation12 applied microarrays on 397 human oocytes and embryos at six developmental stages including fully-grown germinal vesicle oocyte (GV), metaphase I oocyte (MI), metaphase II oocyte (MII), 4-cell embryo (D2), 8-cell embryo (D3) and blastocyst (D5). The authors did not detect any significant differences in expression between the developmental stages of oocytes (GV, MI, MII). Using expression data from all developmental stages, the authors clustered time series of expression levels into 26 expression patterns. The overlap of data from Zhang et al.Citation12 with our H4K12ac ChIP-on-chip indicated that H4K12ac-enriched genes from sperm were represented in 19 of 26 expression clusters in the embryo. The vast majority of H4K12ac-marked genes were not regulated between consecutive stages as shown in pattern 1 (Fig. S4). Twenty-seven genes from anti-H4K12ac immunoprecipitated chromatin were upregulated (2.7%; 27 out of 993 expressed genes) and 41 (3%; 41/1356) genes were downregulated between D2-MII. This data showed that comparable numbers of H4K12ac genes are up- or downregulated in the 4-cell embryo. According to gene ontology classification, H4K12ac-associated genes that are highly expressed in D2 after fertilization are mostly involved in the following processes: gene expression (p value = 1.4e-2), macromolecule metabolic process (p value = 1.4e-2), protein metabolic process (p value = 2.7e-2), RNA metabolic process (p value = 6.2e-2), histone fold (p value = 7.8e-2), transcription DNA dependent (p value = 8.7e-2). This result suggests that particular genes regulating cell division, cell differentiation, transcriptional activity and intracellular metabolism in early embryos are marked with H4K12ac on the paternal genome. Between D3-D2, the upregulation of expression was observed in 23 (3.9%; 23/574) H4K12ac-marked genes, while 18 (3%; 18/586) genes were identified to be downregulated. In the morula stage, H4K12ac activated genes are related to protein metabolic process (p value = 3.0e-4), macromolecular metabolic process (p value = 3.5e-3), cellular protein catabolic process (p value = 7.7e-3), protein modification process (p value = 3.9e-2). Notably, the expression of 39 (2%; 39/1905) H4K12ac-enriched genes was detected at blastocyst stage (D5-D3). Those genes are involved in cellular catabolic process (p value = 9.2E-3), regulation of protein kinase activity (p value = 2.1e-1), cellular developmental process (p value = 4.1e-1) and developmental process (p value = 6.7e-1). The expression of 43 other H4K12ac genes decreased between stages D5-D3 (2.2%; 43/1928). Despite the higher number of H4K12ac promoters, for which gene expression has been found in blastocysts, it is very unlikely that transcriptional regulation in the blastocyst stage is still affected by sperm derived histone modification.
Distribution of H4K12ac in sperm, mouse pronuclei and parthenogenetically activated oocytes
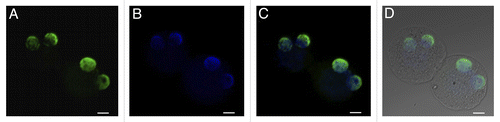
The localization of H4K12ac in human sperm nucleus was analyzed using indirect immunofluorescence. Co-staining of H4K12ac and protamine-1 was conducted using FITC (green) and Cy3 (red) conjugated antibodies, respectively. The signal of H4K12ac was observed in the post-acrosomal region of the sperm head, while protamine-1 was spread over the whole nucleus ().
Figure 8. Distribution of H4K12ac in human sperm nucleus and during mouse early embryo development. Co-localization of H4K12ac (A) and protamine-1 (B) visualized by immunofluorescence using FITC (green) and Cy3 (red) conjugated antibodies (merge) (C). DAPI was used for DNA staining (blue) (D). Immunolabeling of H4K12ac was detected in the post-acrosomal region of the sperm head, while red staining of protamine-1 was spread over the whole nucleus. Immunofluorescent labeling of H4K12ac in early stages of pronuclei establishment (E–H); Pronuclei fusion (I–L); H4K12ac staining (green). Male pronucleus, shown by arrow, gives strong positive signal compares to a smaller female pronucleus shown by asterisk; DNA stained by DAPI (blue) (B,F,); H4K12ac and DAPI (C,G); H4K12ac, DAPI and DIC (D,H). Scale bars represent 10 µm. Photos are merged for each displayed color labeling from several sequenced slices taken by a Leica confocal microscope.
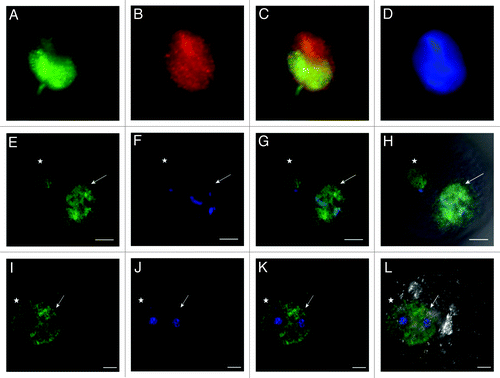
The localization of H4K12ac in mouse fertilized eggs with clearly established pronuclei was analyzed using indirect immunofluorescence. The male and female pronuclei were distinguished by their naturally different size with the male pronucleus being larger than the female pronucleus. There was a strong compact signal over the male pronucleus compared with a weaker one in the female (). During pronuclei migration and at the point of pronuclei fusion, an increasing H4K12ac signal in the female pronucleus could be observed as well, while the male pronucleus remained strongly labeled (). After parthenogenetic oocyte stimulation, both established female pronuclei were positively labeled for H4K12ac (). Presented data represent 10 individual experiments with same results.
Discussion
Considerable attention has already been paid to sperm chromatin packaging, histone to protamine exchange and sperm-derived histone code and its role in early embryonic development.Citation5,Citation6,Citation9,Citation15,Citation16 This study focused on the distribution of a specific histone modification, namely H4K12ac, in human sperm and mouse pronuclei and characterized its specific enrichment sites in promoters throughout the whole human genome.
In a previous study, we reported genome-wide co-localization of H4K12ac binding and endonuclease accessibility indicating that, despite the very different methods of chromatin separation, the CGH and ChIP data sets are substantially equivalent.Citation9 In addition, we know that H4K12ac modified sperm chromatin persists in the zygote,Citation6 and localizes to gene regulatory elements in sperm DNA (CTCF binding sitesCitation9 and promoters of genes involved in developmental processes).Citation14,Citation15
Based on ChIP evaluation with selected promoters, we confirmed the enrichment previously predicted from microarrays; however, strong individual differences were observed between healthy individuals. Furthermore, we analyzed H4K12ac promoter occupancy around TSS and found that binding intensities are symmetrically distributed ± 2 kb from the TSS. Interesting results were obtained from the overlap between anti-H4K12ac binding sites and the sperm mRNA level demonstrating that H4K12ac is associated with particular promoters that may drive high levels of mRNA transcripts stored in mature spermatozoa. The highest expressed mRNA codes for testis specific protein PHF7 (PHD finger protein 7; Testis development protein NYD-SP6; mRNA level 15), suggesting an activating role for H4K12ac in the regulatory elements of this gene. It was also of note that 27 out of 513 promoters associated with H4K12ac in sperm chromatin correlated with genes expressed at 4-cell stage human embryos. Twenty-three H4K12ac-associated genes were activated in 8-cell embryos and 39 genes already in blastocyst. Genes activated in 4-cell embryos are predominantly involved in the control of gene expression, histone folding and DNA-dependent transcription, while genes expressed in the blastocyst stage at day five were classified as involved in developmental processes. The presence of H4K12ac in spermatozoa and its persistence in the zygote was confirmed by immunofluorescent analysis during early mouse embryogenesis, where we followed pronuclei formation, migration and fusion. The difference in H4 acetylation between mouse paternal and maternal chromatin prior to DNA replication was described by Adenot et al.,Citation16 stating that hyperacetylation occurs in paternal, but not maternal, chromatin immediately after fertilization.
In this study, the larger male pronucleus displayed a strong, uniform distribution of H4K12ac. Localization of H4K12ac in the female pronucleus was observed, but with a considerable time delay. Furthermore, during their migration and prior to fusion, the H4K12ac signal in the female pronucleus increased to the same intensity as the male, which remained strongly labeled. Moreover, following parthenogenetic activation of oocytes, both female pronuclei were positively labeled for H4K12ac; thus, an importance of H4K12ac in both pronuclei prior to zygote formation may be inferred. The distribution pattern of H4K12ac in the first cell cycle of in vitro fertilized embryos seems to be conserved between species. According to Maalouf et al.,Citation17 which studied the expression of acetylated lysine residues of histone H4 (K5, 16) in bovine pre-implantation embryos, the signal for H4K12ac in the paternal pronucleus was strong and expanded quickly following pronucleus formation. At the time of fusion, paternal and maternal genomes were equally hyperacetylated. An identical effect has been observed in our experiments with mouse pronucleus, indicating either strong acetylase activity or histones hyperacetylation that has been transmitted from spermatozoa. Interestingly, in parthenogenetically-activated murine oocytes, maternal pronuclei showed high levels of H4K12ac, which was comparable to signals obtained after IVF with sperm. Our results from mice oocytes confirmed the report of Maalouf et al.Citation17 in which the absence of paternal genome leads to a faster enrichment of H4K12ac in parthenots. Considering the fact that parthenogenetically-activated oocytes usually do not reach the 4-cell stage, paternally derived H4K12ac as an activating epigenetic mark may induce the expression of transcription factors and developmentally relevant genes after fertilization.
Carrell and HammoudCitation18 also postulated a hypothetical role for histone modification as a determinant of gene state after fertilization. According to this theory, histone modification promotes either gene activation or repression and, hence, we assume that H4K12ac may be one of those epigenetic marks for developmentally important genes. Comparing the transcriptomic data set from human pre-implantation embryos with H4K12ac occupancy in sperm indicated that only 6% of H4K12ac-associated genes were expressed in 4-cell embryos and 7% in blastocysts. Interestingly, genes expressed in blastocysts are strongly enriched for developmental processes. However, due to chromatin rearrangement during cell division after fertilization, the expression of particular genes could be affected also by maternal histones or newly translated transcription factors. The mechanism of epigenetic regulation in human embryos remains incompletely understood, mainly due to limited accessibility to human material for research purposes. Knowledge about molecular counterparts during embryogenesis comes from studies performed in the murine model. Van der Heijden et al.Citation6 hypothesized that nucleosomal regions provide immediate access for the recruitment of maternal factors that progressively replace protamines in a process that results in the decondensation of paternal chromatin and protamine removal within 30–50 min of gamete fusion. Within an hour after fertilization, the nucleosomal chromatin itself becomes the target of DNA demethylation mechanisms that actively remove the majority of the methylation marks from the paternal genome.Citation19 This process, which is accompanied by histone replacement, is completed within four hours after fertilization. This hypothesis is supported by our observation that, in human sperm, H4K12ac is enriched in regions that are also nucleosomal in murine spermatozoa, and the paternal nucleus acquires maternal H4 histones during decondensation, even though the acquisition of other maternal histones is delayed until after decondensation.
Similar investigations using HG18 NimbleGen array, but with methylated histones, provided new insights into interpretation of paternal histone code in spermatozoa. Results obtained by Bryczynska et al.Citation15 confirmed by the findings by Hammoud et al.Citation14 indicate that H3K4me2 and H3K27me3 are retained at regulatory sequences in mature human spermatozoa. H3K4me2 marks genes that are relevant in spermatogenesis and cellular homeostasis, while H3K27me3 marks developmental regulators in sperm and in somatic cells. Notably, genes with extensive H3K27me3 coverage around transcriptional start sites tend not to be expressed during male and female gametogenesis or in pre-implantation embryos. This finding substantiates the hypothesis of a nucleosomal fraction within developmentally important genes. However, those genes interacting with methylated histone residues seem to be repressed during embryogenesis. Many sperm promoters have been also identified to interact with bivalent histone modifications, such as H3K27me3/H3K4me3. Overlapping the data from CHIP-seq with H3K27me3, H3K4me3, CHIP-on-chip with H3K9ac and H4K12ac, we found very minor co-localization of H4K12ac with methylated histones. This result suggests that H4K12ac may not possess bivalent properties and it is more likely that it is enriched in transcriptionally active promoters.
We also addressed the question of whether there is a preferential distribution of H4K12ac in the sequences near the TSS in sperm chromatin and found that binding intensities are, on the average, highest between 2 kb upstream and downstream the TSS. Brykczynska at al.Citation15 postulated that the probability of paternal epigenetic transmission of epigenetic information by histone modification is expected to depend first on the binding position around the TSS and, second, on the size of the region interacting with the corresponding modification. They also determined the extent to which sequences flanking the TSS contain modified nucleosomes (H3K27me3 and H3K4me2), and this variable was termed “modification coverage.” Computation analysis showed 6,000 bp modification coverage of H3K27me3-marked loci and 3,000 bp of H3K4me2-marked loci. This finding was supported by ontology analyses showing that developmental gene functions are overrepresented among genes that are more broadly marked by H3K27me3 in spermatozoa.
The relationship between H4K12ac promoter occupancy and mRNA level with respect to genes stored in spermatozoa, as well as its protein localization in paternal chromatin, are another important result that has to be taken into consideration. In promoters with high enrichment of H4K12ac, we detected high mRNA level stored in spermatozoa.Citation20 Assuming that sperm cells are transcriptionally inactive cells, the presence of mRNA transcripts in mature spermatozoa probably reflects transcriptional activity during spermatogenesis. Based on our observations, we could speculate that acetylation of histones and in particular H4K12ac may contribute to activation of certain genes prior to their transcriptional arrest. Previous theories that mRNAs are left over from spermatogenesis were valid until it was discovered that spermatozoa deliver a unique set of mRNAs to the oocyte at fertilization as a prerequisite for proper embryogenesis.Citation21-Citation24 Furthermore, the two mechanisms, H4K12ac interaction at specific promoters and expression of the corresponding mRNA transcripts, may represent a cascade of events resulting in transition of developmentally important mRNA transcripts to the zygote. In cases of male infertility with altered histone retention, aberrant mRNA profiles may also be expected. In preliminary studies with subfertile patients presenting aberrant chromatin condensation as assessed by aniline staining, we found a global reduction of H3K9ac marked sites.Citation13 However, mRNA profiling has not been yet studied in these individuals, although our studies related to epigenetic modifications of bromodomain testis-specific protein (BRDT) in spermatozoa indicate reduced enrichment of methylated histones and increased mRNA level of BRDT in subfertile patients.Citation25 Data were compared with the clinical outcomes of patients that correlated barely in aniline blue staining and sperm morphology. This is in agreement with mRNA fingerprints differing substantially between normospermic and teratozoospermic men.Citation26,Citation27 These findings support the idea that aberrant histone modifications may be associated with impaired expression of developmentally important mRNA transcripts in teratozoospermic men, which may negatively influence fertilization rate and, ultimately, affect embryogenesis.
Material and Methods
ChIP-on-chip
Chromatin immunoprecipitation (ChIP) was performed following the protocol provided by UpstateMillipore at http://www.millipore.com. However, several modifications were necessary due to the fact that only an estimated 10–15% of the human sperm genome remains in a nucleosomal context as result of the histone to protamine replacement during spermatogenesis.Citation8
Briefly, 1 x 107 sperm cells were used per immunoprecipitation with antibodies. After the collection of ejaculates from healthy donors, sperm cells were separated from seminal plasma and washed twice in 1x Dulbecco’s PBS by 5 min centrifugation at 300 x g. In this study we performed ChIP-chip in three replicates from freshly obtained ejaculate from healthy donors. The swim–up procedure allowed us the separation of motile sperm from immotile cell fraction. In addition, applying of lysis buffer (1% SDS, 10mM EDTA, 50 mM TRIS-HCl, pH 8.0) for 5 min in initial steps of CHIP assay enables breaking out of somatic cells instead of sperm as sperm cells are much robust and usually require a mechanistic disrupter with homogenizer. The lysis of round cells is normally monitored under the standard light microscopy. Spermatozoa were subsequently washed 2 times with washing buffer and again treated with Lysis buffer for 20 min and 20 strokes in an Ultra Turrax homogenizer were performed for separation of sperm heads from sperm tails. To rule out any contamination on the level of isolated DNA, we checked sonicated DNA in respect of somatic mRNA presence in using Experion (Bio-Rad) in sample without RNase treatment and purified RNA. The peak of rRNA could not be detected indicating lack of contamination of somatic fraction.
ChIP procedure was performed as previously described by Steilmann et al.Citation13 In this study we used polyclonal IgG ChIP grade (ab46540, Abcam) as a negative control and antibodies against unmodified histone H3 (ab1791, Abcam) as a positive control. 10% of sonicated chromatin (100 µl) was saved for each sample to determine the input chromatin amount. ChIP grade rabbit polyclonal antibodies against acetylated histone H4 at lysine 12 (H4K12ac) used for immunoprecipitation were purchased from Abcam (ab 1761). After addition of 40 µl of salmon sperm, DNA was extracted by phenol/chloroform treatment, ethanol precipitation with 3 µl glycogen as inert carrier, resuspended in 30 µl of nuclease free water analyzed by qPCR or in combination with microarray. For microarray (chip) analysis, immunoprecipitated (IP) sample was purified with Qiaquick Purification Kit (Qiagen) and 10 ng of input material (total chromatin) was prepared by adapting the protocol for whole genome amplification using the Sigma GenomePlex WGA kit (http://www.sigmaldrich.com) as described in O’Green et al.Citation28 Initial random amplification step of WGA protocol has been omitted due to previous defragmentation of sperm chromatin after sonication. The required amount of DNA for microarray analysis (4 µg per sample) was generated using WGA Re-amplification Kit (Sigma-Aldrich, Steinheim, Germany). Amplicons were applied to the HG18 human 5 kb promoter array from NimbleGen System (see http://www.nimblegen.com for details). The 5 kb-promoter array set consists of two individual arrays with a total of approximately 24.000 50mer gene promoter region probes with spacing of about 100 bp. Array platform 1 covered gene promoters localized on chromosomes 1–10 while array platform 2 covered gene promoters localized on chromosomes 11-XY. Fluorescence signal intensity data were extracted from the scanned images of each array using data extraction software. Each feature on the array has a corresponding scaled log2-ratio that was calculated from the input signal-Cy3 for the total chromatin and IP-probe –Cy5, which were co-hybridized to the array. The log2 ratio was computed and scaled to the center the ratio data around zero. Scaling is performed by subtracting the bi-weight mean for the log2-ratio values for all features on the array from each log2-ratio value. This data analysis provided by NimbleGen is equivalent to a mean normalization of each channel. Binding sites for H4K12ac to sperm chromatin was detected by searching for 4 or more oligo probes whose signals were above the specific cutoff values, ranging from 90% to 15%, using a 500 bp sliding window. The ratio data were randomized 20 times and each peak was assigned a false discovery rate score (FDR) based on the randomization. The lower the FDR score, the more likely the peak corresponded to H4K12ac binding site. Data report from ChIP-on-chip assay contained ranking list of all identified peaks with an FDR < 0.2 with the annotation to each gene promoter as accessed using the known gene database available at http://www.ncbi.nlm.nih.gov/entrez/query.fcgi.
H4K12ac ChIP assay quality assessment and averaging of replicates
The population of ejaculated spermatozoa from healthy men contains a certain level of heterogeneity concerning the fertilization capacity. The routine ejaculate diagnostic provides evidence of the variability between healthy individuals, although those men exhibit all normal sperm parameters, according to the WHO guidelines. Three different Chip-on-chip replicates were hybridized using ejaculates from a healthy donor and a control. The Bioconductor package arrayQualityMetrics was applied for visualization and metrics for assessing microarray data quality.Citation29 In our ChIP-on-chip set, the Cy3 green channel holds the intensities from the untreated input sample and the Cy5 channel holds the immunoprecipitate of H4K12ac. When comparing intensity correlations between replicates, higher values could be identified between replicate 1 and the biological replicate (Spearmann Correlation 0.783) (Fig. Nine A). Replicates from donor 1 and donor 2 were kept for further analysis by averaging and mapping enriched reporters to the genome, while technical replicates were excluded. Raw data that we used for this study have been submitted to NCBI Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo/) under accession nos. GSM406528–GSM406531.
Finding H4K12ac enriched regions in sperm
In order to find H4K12ac enriched regions in sperm DNA, we applied the Ringo package, which estimates a mixture of two underlying distributions. One is the null distribution of reporter levels in non-enriched regions; the other is the alternative distribution of the levels in enriched regions. A critical parameter in algorithms for the detection of ChIP-enriched regions is the fraction of reporters on the array that is expected to show enrichment.Citation30
Results are shown as an enrichment histogram () demonstrating mean signal ratios of H4K12ac chromatin-immunoprecipitated DNA vs. random-sheared total genomic DNA (Input). The distinct tail at the right-hand end corresponds to DNA fragments enriched by H4K12ac. Tiled oligos that displayed the top 3% ratios are located to the right of the red bar. The level of enrichment of each position is reflected as an area under the curve score that is, the sum of the smoothed reporter levels, each minus the threshold. Regions with high ChIP-on-chip scores are more likely to have the motifs of interest.Citation31 Using these criteria, we identified 514 gene promoters as potential binding sites for H4K12ac. The promoter for PEX26 on chromosome 22 was identified as the highest peak with the score of 5.2 and maximum level 2.6 after averaging and normalization of our data (; Fig.S1).
Validation of enriched promoters with ChIP in combination with real-time polymerase chain reaction (PCR)
ChIP assays were validated by quantitative real-time PCR. Primers for enriched sequences including 1kb upstream and downstream from the peak center were generated. Primers are listed in . Additionally we performed ChIP assays with newly available chip-grade antibodies against protamine-1 (Catalog no. mAb-001, Briar Patch Biosciences). PCR reactions contained 2 μl of immunoprecipitation sample, 10 pmol/μl of each primer, 12.5 μl iQTM SYBR Green Supermix (Catalog no.170–8882, Bio-Rad) in 23 μl of total volume. The PCR conditions were an initial step of 3 min. at 95°C, followed by 40 cycles of 30 sec. at 95°C, 30 sec. at 58–62°C and 1 min. at 72°C. Furthermore, PCR products were checked by sequencing (Scientific Research and Development GmbH).
Table 1. Primer pairs used for validation of H4K12ac binding with real-time qPCR
Calculations for graphs were processed as follows: Ct is the cycle number at which each PCR reaction reaches a predetermined fluorescence threshold. It is set within the log linear range of all reactions. Enrichment of the immunoprecipitated sample compared with input material was calculated as follows: ΔCt = Ct (input) - Ct (immunoprecipitated sample) and % total = 2ΔCt × 10 (according to 10% input chromatin of total immunoprecipitated chromatin).Citation32 For fertile patients, PCRs with material of different donors (n = 3) were performed. The mean of the Ct values of different donors was calculated.
Comparative genomics: Data set of genes associated with H4K12ac in sperm with expression profiling of mRNAs from human sperm of fertile donors
In order to investigate whether promoter association with H4K12ac in specific promoters is possibly correlated with mRNA transcripts stored in mature spermatozoa of fertile men, we performed comparative analysis of data set of genes interacting with H4K12ac in sperm (Chromatin Immunoprecipitation with H4K12ac in combination with promoter microarray HG18 NimbleGen) with genome wide expression profiling of mRNAs from human sperm of fertile men. (CodeLink Human Whole Genome Bioarray, Applied Microarrays). Twelve human ejaculates from patients who underwent fertility treatment and fertilization resulted in pregnancy were used in this study. Ejaculates were liquefied for 30 min at 37°C and an aliquot of 1 mL was washed once by centrifugation in PBS (500 g). Total RNA was extracted from the ejaculates by a specially designed 2-fold genomic DNA pre-elimination protocol.Citation32 50 - 500 ng total RNA were amplified with the MessageAmp III Kit (Catalo no. AM1794, Ambion) and Biotin-UTP was incorporated as a label. Hybridization was conducted for 20h at 37°C and arrays were washed in 1x TNT (10 min, room temperature) and 1x TNT (60 min, 46°C). Staining with Cy5-Streptavidin was performed for 30 min at room temperature. Arrays were then washed 2 times with 1x TNT for 10 min and 0.2x SSPE for 30s and spin-dried in a centrifuge at 2000 rpm for 2 min.
Scanning was done on a ScanArray Gx Scanner (Perkin Elmer) at PMT 70/LP 90% setting and 5 µm resolution. Images were acquired with the ScanArray Express 3.0 software (Perkin Elmer). Scan images were converted to raw values by the CodeLink Expression analysis software (GE Healthcare). Genes with lower than 5th percentile expression values over all samples were removed. Normalization was by quantile normalization using the Bioconducter package affy. Detailed information and data sets are to be found under Gene Expression Omnibus GEO Accession: GSE29002. (www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=hngtryockgwcalu&acc=GSE29002). Matching of chromatin Immunoprecipitation/promoter microarray data with sperm gene expression data was based on intersecting gene names (HUGO ordinary gene symbols).
Immunofluorescence of human sperm
Ejaculated sperm from healthy donors were washed three times with Tris buffered saline (TBS) and 10µl of the sperm suspension was smeared onto a glass slides. Samples were then dried at room temperature for 2 h. To facilitate antibody penetration into sperm nucleus, cells were treated with buffer containing DTT and heparin, which caused decondensation of sperm chromatin. However, we have observed that the incubation with decondensing buffer was stopped after max. Fifteen min, as longer treatment would cause damage to the physiological histone-protamine compartment in the sperm. Prior to incubation with antibodies, sperm cells were treated with 100µl of freshly prepared decondensing mix (25 mM dithiothreitol, 0.2% Triton- X-100, 200 IU heparin/ml in PBS) for 20 min to improve the accessibility of nuclear proteins organized in strongly compacted sperm chromatin.Citation6 Next, slides were fixed using 4% paraformaldehyde in PBS (pH 7.0) for 15 min, followed by incubation with 10% SDS for 25 min. After the washing steps, slides were incubated with 5% BSA in Tris buffered saline (TRIS-HCl, pH 7.4) for 2 h in a humidified chamber. Then a co-incubation with two secondary antibodies against protamine-1 [goat polyclonal antibody (N-14), Catalog no. sc-23105, Santa Cruz Biotechnology; dilution 1:200] and against acetylated histone H4 at lysine 12 (H4K12ac) (rabbit polyclonal antibody, Catalog no. ab1706, Abcam; dilution 1:20) was performed at 4°C overnight. Fluorescein thiocyanate FITC-conjugated secondary antibody goat anti-rabbit (Catalog no. ab97050, Abcam) and cyanine derivative Cy3 conjugated donkey anti-goat (catalog no. ab6949, Abcam) were added to the reaction in the dilution 1:50 and 1:400, respectively, and incubated for 1 h at 37°C in a humidified chamber. After washing with TBS for 5 min, slides were mounted with one drop of Faramount Mounting Medium containing DAPI (Order no. S302580, DAKO). The stained cells were visualized with a Zeiss Cell observer inverse epifluorescence microscope system and processed with Adobe Photoshop CS2 Version 9.
Mouse in vitro fertilization and immunostaining of mouse pronuclei and parthenogenetically activated oocytes
C57Bl/6 mice were obtained from a breeding colony of the Laboratory of Reproduction, Faculty of Science, Charles University in Prague. Female mice used for hormonal stimulation were 21 d old and male mice were in a reproductive age of 10 to 12 weeks. All animal procedures were performed in strict accordance with the Animal Scientific Procedure, Art 2010, and subjected to review by the local ethics committee.
Eggs were isolated from oviduct of juvenile hormonally stimulated females (strain C57Bl/6) and added to capacitating sperm obtained from distal region of cauda epididymis. After in vitro fertilization, the zona pellucida was removed by Tyrodes solution (Catalog no. T1788, Sigma) and eggs were fixed for 20 min in 2.5% paraformadehyde, washed and permeabilized for 10 min by 0.1% TritonX-100 in PBS. For immunofluorescent labeling, eggs were blocked in 5% goat serum in PBS, for 1h, followed by primary polyclonal antibody anti-H4K12ac, (ab1761, Abcam), dilution 1:200 in 1% goat serum for 2 h. After washing, a secondary anti-rabbit polyclonal antibody (ab6717, Abcam, dilution 1:1000 in PBS) was added for 1 h. Slides were mounted into Vectashield mounting medium with DAPI (Catalog no. H-1200, Vector Lab). Samples were examined with a Leica DM IRE2 High-speed confocal/two photon system for Live Cell Imaging and Dynamics (Czech Republic).
| Abbreviations: | ||
| AFF4 | = | AF4/FMR2 family member 4 |
| AGPAT6 | = | Glycerol-3-phosphate acyltransferase 4 precursor |
| AXIN1 | = | axis inhibition protein 1 |
| BRDT | = | bromodomain testis specific protein |
| CBX8 | = | chromobox protein homolog 8 |
| CHIP | = | chromatin immunoprecipitation assay |
| CHIP-on-chip | = | chromatin immunoprecipitation assay in combination with microarray |
| CTCF | = | CCCTC- binding factor |
| Cy3 | = | cyanine 3 conjugate |
| D2 | = | day 2, 4-cell embryo |
| D3 | = | day 3, 8-cell embryo |
| D5 | = | day 5 blastocyst |
| DAPI | = | 4',6-diamidino-2-phenylindole fluorescent nucleic acids stain |
| DIC | = | differential interference contrast |
| DVL1 | = | segment polarity protein disheveled homolog DVL-1 |
| EP300 | = | histone acetyltransferase p300 |
| FDR | = | false discovery rate |
| FITC | = | fluorescein thiocyanate |
| FZD3 | = | frizzled-3 precursor |
| GMCL1 | = | germ cell-less protein-like 1 |
| GV | = | germinal vesicle oocyte |
| H3K27me3 | = | histone H3 trimethylated at lysine 27 |
| H3K4me3 | = | histone H3 trimethylated at lysine 4 |
| H3K9ac | = | histone H3 acetylated at lysine 9 |
| H4K12ac | = | histone H4 acetylated at lysine 12 |
| HDAC6 | = | histone deacetylase 6 |
| HIST1H2BC | = | histone H2B type 1-C/E/F/G/I |
| HSF2BP | = | heat shock factor 2-binding protein |
| IFT172 | = | intraflagellar transport protein 172 homolog |
| ITCH | = | myosin light chain 3 |
| KDM2A | = | lysine-specific demethylase 2A |
| MAPK8IP3 | = | C-jun-amino-terminal kinase-interacting protein 3 |
| MAST2 | = | microtubule-associated serine/threonine-protein kinase 2 |
| MI | = | metaphase I oocyte |
| MII | = | metaphase II oocyte |
| MLH3 | = | DNA mismatch repair protein Mlh3 |
| NCOA6 | = | nuclear receptor co-activator 6 |
| NSD1 | = | histone-lysine N-methyltransferase |
| PEX9 | = | peroxisomal membrane protein 9 |
| PHF7 | = | testis-specific PHD finger protein-7 |
| PSMC4 | = | 26S protease regulatory subunit 6B |
| RUVBL1 | = | U6 spliceosomal RNA |
| STAG3 | = | cohesin subunit SA-3 (stromal antigen 3) |
| TCFL5 | = | transcription factor-like 5 protein |
| TRIP13 | = | thyroid receptor-interacting protein 13 |
| TSS | = | transcription star site |
| USP9X | = | probable ubiquitin carboxyl-terminal hydrolase FAF-X |
| WHO | = | World Health Organization |
| ZEB1 | = | zinc finger E-box-binding homeobox 1 |
Additional material
Download Zip (450.7 KB)Acknowledgments
Financial support for this study was available from a grant of the German Research Foundation (DFG), Project 1 of the Clinical Research Unit KFO 181/2.
The part of the work concerning mouse embryos was supported by Institutional Research Support grant No. SVV-2012–265 206 and by the Grant Agency of the Czech Republic GACR No. P506/12/1046. We thank Angela Erkel and Barbara Fröhlich for technical support and Farhad Asskaryar for critical reading.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental Material
Supplemental materials may be found here: www.landesbioscience.com/journals/epigenetics/article/21556
References
- Björndahl L, Kvist U. Human sperm chromatin stabilization: a proposed model including zinc bridges. Mol Hum Reprod 2010; 16:23 - 9; http://dx.doi.org/10.1093/molehr/gap099; PMID: 19933313
- Ylij JMG, Schmidz W, Morton E. Zinc-induced Protamines ” Secondary Structure Transitions in Human Sperm. Peptides 1990; 265:20667 - 72
- Kimmins S, Sassone-Corsi P. Chromatin remodelling and epigenetic features of germ cells. Nature 2005; 434:583 - 9; http://dx.doi.org/10.1038/nature03368; PMID: 15800613
- Lachner M, Jenuwein T. The many faces of histone lysine methylation. Curr Opin Cell Biol 2002; 14:286 - 98; http://dx.doi.org/10.1016/S0955-0674(02)00335-6; PMID: 12067650
- van der Heijden GW, Dieker JW, Derijck AA, Muller S, Berden JH, Braat DD, et al. Asymmetry in histone H3 variants and lysine methylation between paternal and maternal chromatin of the early mouse zygote. Mech Dev 2005; 122:1008 - 22; http://dx.doi.org/10.1016/j.mod.2005.04.009; PMID: 15922569
- van der Heijden GW, Derijck AA, Ramos L, Giele M, van der Vlag J, de Boer P. Transmission of modified nucleosomes from the mouse male germline to the zygote and subsequent remodeling of paternal chromatin. Dev Biol 2006; 298:458 - 69; http://dx.doi.org/10.1016/j.ydbio.2006.06.051; PMID: 16887113
- Ajduk A, Yamauchi Y, Ward MA. Sperm chromatin remodeling after intracytoplasmic sperm injection differs from that of in vitro fertilization. Biol Reprod 2006; 75:442 - 51; http://dx.doi.org/10.1095/biolreprod.106.053223; PMID: 16775225
- van der Heijden GW, Ramos L, Baart EB, van den Berg IM, Derijck AA, van der Vlag J, et al. Sperm-derived histones contribute to zygotic chromatin in humans. BMC Dev Biol 2008; 8:34; http://dx.doi.org/10.1186/1471-213X-8-34; PMID: 18377649
- Arpanahi A, Brinkworth M, Iles D, Krawetz SA, Paradowska A, Platts AE, et al. Endonuclease-sensitive regions of human spermatozoal chromatin are highly enriched in promoter and CTCF binding sequences. Genome Res 2009; 19:1338 - 49; http://dx.doi.org/10.1101/gr.094953.109; PMID: 19584098
- Urano A, Endoh M, Wada T, Morikawa Y, Itoh M, Kataoka Y, et al. Infertility with defective spermiogenesis in mice lacking AF5q31, the target of chromosomal translocation in human infant leukemia. Mol Cell Biol 2005; 25:6834 - 45; http://dx.doi.org/10.1128/MCB.25.15.6834-6845.2005; PMID: 16024815
- Yamauchi T, Yamauchi J, Kuwata T, Tamura T, Yamashita T, Bae N, et al. Distinct but overlapping roles of histone acetylase PCAF and of the closely related PCAF-B/GCN5 in mouse embryogenesis. Proc Natl Acad Sci U S A 2000; 97:11303 - 6; http://dx.doi.org/10.1073/pnas.97.21.11303; PMID: 11027331
- Zhang P, Zucchelli M, Bruce S, Hambiliki F, Stavreus-Evers A, Levkov L, et al. Transcriptome profiling of human pre-implantation development. PLoS One 2009; 4:e7844; http://dx.doi.org/10.1371/journal.pone.0007844; PMID: 19924284
- Steilmann C, Paradowska A, Bartkuhn M, Vieweg M, Schuppe HC, Bergmann M, et al. Presence of histone H3 acetylated at lysine 9 in male germ cells and its distribution pattern in the genome of human spermatozoa. Reprod Fertil Dev 2011; 23:997 - 1011; http://dx.doi.org/10.1071/RD10197; PMID: 22127005
- Hammoud SS, Nix DA, Zhang H, Purwar J, Carrell DT, Cairns BR. Distinctive chromatin in human sperm packages genes for embryo development. Nature 2009; 460:473 - 8; PMID: 19525931
- Brykczynska U, Hisano M, Erkek S, Ramos L, Oakeley EJ, Roloff TC, et al. Repressive and active histone methylation mark distinct promoters in human and mouse spermatozoa. Nat Struct Mol Biol 2010; 17:679 - 87; http://dx.doi.org/10.1038/nsmb.1821; PMID: 20473313
- Adenot PG, Mercier Y, Renard JP, Thompson EM. Differential H4 acetylation of paternal and maternal chromatin precedes DNA replication and differential transcriptional activity in pronuclei of 1-cell mouse embryos. Development 1997; 124:4615 - 25; PMID: 9409678
- Adenot PG, Mercier Y, Renard JP, Thompson EM. Differential H4 acetylation of paternal and maternal chromatin precedes DNA replication and differential transcriptional activity in pronuclei of 1-cell mouse embryos. Development 1997; 124:4615 - 25; PMID: 9409678
- Carrell DT, Hammoud SS. The human sperm epigenome and its potential role in embryonic development. Mol Hum Reprod 2010; 16:37 - 47; http://dx.doi.org/10.1093/molehr/gap090; PMID: 19906823
- Santos F, Hendrich B, Reik W, Dean W. Dynamic reprogramming of DNA methylation in the early mouse embryo. Dev Biol 2002; 241:172 - 82; http://dx.doi.org/10.1006/dbio.2001.0501; PMID: 11784103
- Nanassy L, Carrell DT. Paternal effects on early embryogenesis. J Exp Clin Assist Reprod 2008; 5:2; http://dx.doi.org/10.1186/1743-1050-5-2; PMID: 18485208
- Ostermeier GC, Dix DJ, Miller D, Khatri P, Krawetz SA. Spermatozoal RNA profiles of normal fertile men. Lancet 2002; 360:772 - 7; http://dx.doi.org/10.1016/S0140-6736(02)09899-9; PMID: 12241836
- Ostermeier GC, Goodrich RJ, Moldenhauer JS, Diamond MP, Krawetz SA. A suite of novel human spermatozoal RNAs. J Androl 2005; 26:70 - 4; PMID: 15611569
- Miller D, Ostermeier GC. Towards a better understanding of RNA carriage by ejaculate spermatozoa. Hum Reprod Update 2006; 12:757 - 67; http://dx.doi.org/10.1093/humupd/dml037; PMID: 16882702
- Boerke A, Dieleman SJ, Gadella BM. A possible role for sperm RNA in early embryo development. Theriogenology 2007; 68:Suppl 1 S147 - 55; http://dx.doi.org/10.1016/j.theriogenology.2007.05.058; PMID: 17583784
- Steilmann C, Cavalcanti MCO, Bartkuhn M, Pons-Kühnemann J, Schuppe H-C, Weidner W, et al. The interaction of modified histones with the bromodomain testis-specific (BRDT) gene and its mRNA level in sperm of fertile donors and subfertile men. Reproduction 2010; 140:435 - 43; http://dx.doi.org/10.1530/REP-10-0139; PMID: 20538714
- Platts AE, Dix DJ, Chemes HE, Thompson KE, Goodrich R, Rockett JC, et al. Success and failure in human spermatogenesis as revealed by teratozoospermic RNAs. Hum Mol Genet 2007; 16:763 - 73; http://dx.doi.org/10.1093/hmg/ddm012; PMID: 17327269
- Rawe VY, Díaz ES, Abdelmassih R, Wójcik C, Morales P, Sutovsky P, et al. The role of sperm proteasomes during sperm aster formation and early zygote development: implications for fertilization failure in humans. Hum Reprod 2008; 23:573 - 80; http://dx.doi.org/10.1093/humrep/dem385; PMID: 18089554
- Geen HO, Nicolet CM, Blahnik K, Green R, Peggy J. NIH Public Access. Culture 2008; 41:577 - 80
- Toedling J, Huber W. Analyzing ChIP-chip data using bioconductor. PLoS Comput Biol 2008; 4:e1000227; http://dx.doi.org/10.1371/journal.pcbi.1000227; PMID: 19043553
- Schwartz YB, Kahn TG, Nix DA, Li XY, Bourgon R, Biggin M, et al. Genome-wide analysis of Polycomb targets in Drosophila melanogaster. Nat Genet 2006; 38:700 - 5; http://dx.doi.org/10.1038/ng1817; PMID: 16732288
- Shim H, Keles S. Integrating quantitative information from ChIP-chip experiments into motif finding. Biostatistics 2008; 9:51 - 65; http://dx.doi.org/10.1093/biostatistics/kxm014; PMID: 17533175
- Frank SR, Schroeder M, Fernandez P, Taubert S, Amati B. Binding of c-Myc to chromatin mediates mitogen-induced acetylation of histone H4 and gene activation. Genes Dev 2001; 15:2069 - 82; http://dx.doi.org/10.1101/gad.906601; PMID: 11511539