Abstract
Histone deacetylases (HDACs) play a crucial role in chromatin structure and, consequently, gene expression. Their deregulation has been reported in various cancers. We performed a complete and comprehensive study of the expression of 18 HDACs (including Sirtuin; SIRT) by real-time PCR in a cohort of 200 chronic lymphocytic leukemia (CLL) patients with a median follow-up of 77 mo, and compared it with the results obtained from normal B cells. We also compared HDAC expression at diagnosis and after relapse. We observed significant deregulation (mostly upregulation) of HDACs in CLL. In terms of clinical significance, only HDAC6 was significantly correlated with treatment-free survival (TFS), whereas HDAC3 and SIRT2, 3 and 6 were correlated with overall survival (OS). A multivariate Cox regression stepwise analysis indicated that HDAC6, 7 and 10 and SIRT3 were TFS independent predictors. Interestingly, poor prognosis was associated with an overexpression of HDAC7 and 10 but an underexpression of HDAC6 and SIRT3. Therefore, these factors were combined in a TFS score: patients with a score of 0–1–2, 3 and 4 had a median TFS of 107, 57 and 26 mo, respectively (HR = 4.03, p < 0.0001). For OS, SIRT5 and 6 allowed stratification into 3 groups, with a median OS of > 360, 237 and 94 mo (HR = 6.38, p < 0.0001). However, we could not find statistical differences in HDAC expression after relapse. These results, validated by a 5-fold cross-validation, highlight the complex impact of HDAC expression in CLL clinical course.
Introduction
Epigenetic histone modifications play a crucial role in chromatin structure. Among enzymes regulating these processes, histone deacetylases (HDACs) can remove acetyl groups from histone tails, thus increasing their interaction with DNA and leading to chromatin condensation.Citation1,Citation2 HDAC can consequently regulate gene expression by modifying epigenetic configuration in both transformed and non-transformed cells.Citation3 To date, the HDAC family is composed of 18 isoforms in humans, which can be classified into 4 classes based on their structure and cellular localizationCitation4: class I (HDAC1, 2, 3 and 8) and class II (HDAC4, 6, 7, 9,and 10) are the most studied, whereas little is known about class III [Sirtuin (SIRT) 1 to 7] and class IV, which comprises only a single member (HDAC11). Recently, these enzymes have attracted increased interest because HDAC inhibitors (HDACis) have been considered as new and promising therapeutic agents for the treatment of solid cancers and hematological malignancies.Citation5 In addition, deregulation of HDAC expression has been observed in several cancer types, such as breast, lung, ovarian, prostate cancer and lymphoma, highlighting the importance of epigenetics in tumoral development (reviewed in ref. Citation6). However, the relationship between HDAC expression and clinicopathological characteristics in different cancer types remains controversial and complex: the overexpression of a specific HDAC could be associated with a favorable prognosis in one cancer but with poor prognosis in another.Citation6 An individual and comprehensive study of HDAC expression is thus needed for each cancer type.
Chronic lymphocytic leukemia (CLL) is a disease characterized by a clonal accumulation of neoplastic B cells. This leukemia displays two types of prognoses:Citation7 some patients rapidly progress and die, while others remain asymptomatic for many years. This clinical evolution can currently be predicted by several prognostic factors, such as ζ- associated protein-70 (ZAP70), lipoprotein lipase (LPL) and CD38 molecule (CD38);Citation8 CLL is, up to now, incurable, despite the introduction of new treatments. In vitro studies have shown that HDAC inhibitors, such as valproic acid (VPA)Citation9 and suberoylanilide hydroxamic acid (SAHA),Citation10 can induce apoptosis of CLL cells, and clinical trials combining HDACis and classical chemotherapy are currently ongoing.Citation11 However, a comprehensive and complete study of HDAC expression in CLL is missing. Indeed, to our knowledge, no study has correlated HDAC expression with the clinical evolution of CLL patients.
In the present study, we aimed to determine the mRNA expression pattern of the 18 human HDAC isoforms by real-time PCR in CD19+ purified B cells isolated from a clinically well-characterized CLL patient cohort and compare them with 20 normal CD19+ purified B cell samples obtained from peripheral blood of healthy donors and CD19+ purified B cells isolated from 20 CD5+ cord blood samples. These expressions levels were subsequently correlated with classical and well-known prognostic factors, treatment-free survival (TFS) and overall survival (OS) in our cohort of 200 CLL patients, with a median follow-up of 77 mo.
Results
HDAC expression is deregulated in CLL compared with normal B cells
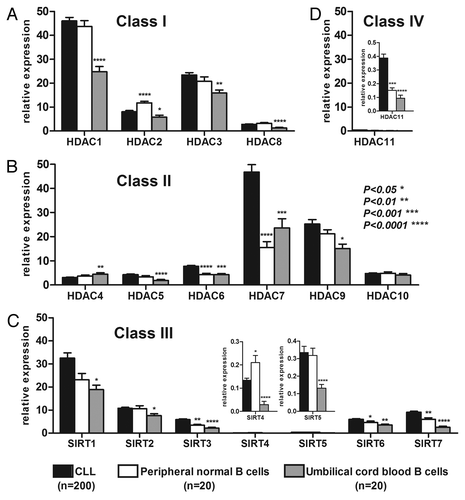
As presented in , HDAC expression was variable from one isoenzyme to another: although HDAC-1 and -7 were highly expressed in CLL, HDAC11, SIRT4 and SIRT5 levels were low. These expressions levels were compared with those of normal B cells purified from the peripheral blood of healthy donors (PB) and purified B cells from umbilical cord blood (UCB) as controls. UCB was chosen as the second control because these B cell samples were CD5+, which is similar to the CLL samples. Compared with PB B cells, HDAC2 and SIRT4 were significantly downregulated, whereas HDAC-6, -7 and -11 and SIRT-3, -6 and -7 were upregulated in CLL samples (p < 0.05). Compared with UCB B cells, all of the isoenzymes were significantly upregulated in CLL samples except HDAC4, which was downregulated, and HDAC10, which was not significantly different (Table S1). These results demonstrated that HDAC expression was deregulated (mostly upregulated) in CLL compared with normal B controls.
Figure 1. HDAC expression in CLL, PB and UCB samples. The mean expression of the different HDACs in CLL (n = 200), PB (n = 20) and UCB (n = 20) was plotted with the standard error of the mean (SEM) according to HDAC class I (A), class II (B), class III (C) and class IV (D). HDAC values are expressed as relative fold change normalized to cyclophilin gene expression and calibrated with a common value. Significant differences were assessed using the Mann-Whitney non-parametric test. Statistical details can be found in Table S1.
Association of HDAC expression with classical prognostic factors and TFS/OS
We compared HDAC expression in different prognosis subgroups based on classical prognostic factors. As shown in Table S1, there were sparing significant differences between some prognostic subgroups. However, the fold changes of HDAC expression had a small amplitude, from -1.5 to +1.6, and no global conclusion could be reached. However, for the majority of HDACs, patients who died generally had lower HDAC expression, and these differences were significant for HDAC-1, -3, -6 and SIRT-2 and -3.
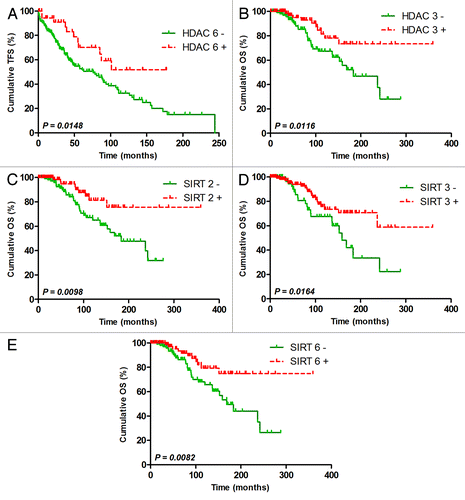
Using ROC curve analysis to maximize the concordance with the ZAP70 status and minimize the number of false negatives, we defined HDAC cut-offs. Although these cut-offs were not optimized for TFS/OS prediction, they were sufficient to observe the following significant differences: taken individually, only HDAC6 was a significant TFS predictor, while HDAC3 and SIRT-2, -3 and -6 could significantly predict OS (). Interestingly, all of the good prognosis subgroups were characterized by a high expression of these HDACs, indicating that low HDAC expression could be associated with unfavorable prognosis. Although the results obtained for HDAC7 (p = 0.0971) and HDAC10 (p = 0.1066) were not significant, we observed that their higher expression could be associated with a poor prognosis, suggesting that no general conclusions associating HDAC expression with prognosis could be drawn. Kaplan-Meier curves and the median TFS/OS for all HDAC subgroups are shown in Figure S1 (TFS), Figure S2 (OS) and Table S3.
Figure 2. Significant TFS or OS power of HDAC expression. Representative TFS curves for HDAC6 (A) and OS curves for HDAC3 (B), SIRT2 (C), SIRT3 (D) and SIRT6 (E). HDACs were measured by real-time PCR, and cut-offs were optimized to maximize ZAP70 concordance and minimize false negatives using ROC curve analyses. Significant differences between curves were calculated using the log-rank test. Statistical details can be found Table S3.
Univariate and multivariate Cox analysis and HDAC score construction and validation
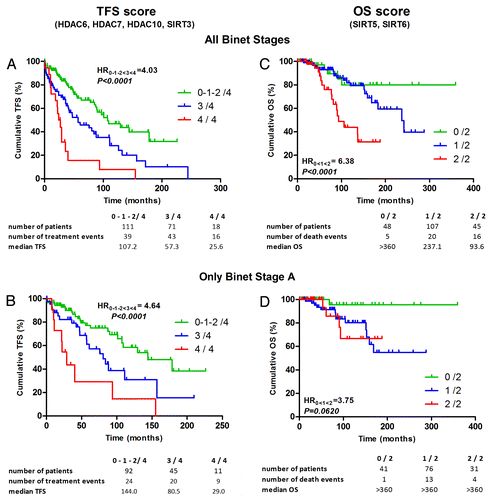
Univariate Cox analysis confirmed the Kaplan-Meier results (). Because treatment and survival appeared to be influenced by several HDACs, we performed a multivariate stepwise Cox regression model to evaluate the impact of all dichotomized HDACs (using Table S3cut-offs) on TFS and OS. Interestingly, for TFS prediction, 4 HDACs were selected: HDAC-6, -7, -10 and SIRT3 (). However, only HDAC6 was a significant TFS predictor in univariate Cox analysis. Regarding the hazard ratios (HR), we observed that HDAC7 and 10 were higher than 1, indicating that the patient had an increasing chance to be treated when these HDACs were highly expressed. In contrast, HDAC6 and SIRT3 displayed an HR < 1, indicating that a positive status for these HDACs is associated with a good prognosis. We then generated a TFS score based on these four HDACs. This score varied from 0 to 4, according to the number of unfavorable factors (i.e., low expression of HDAC6 or SIRT3 and high expression of HDAC-7 or -10), and it was applied to our 200-patient cohort. The presence of a poor prognostic marker corresponds to an increase of 1 unit in the final TFS score. We thus gave the same weight to all four factors. According to this score, the patients were thus stratified into three groups (0–1–2/4, 3/4 and 4/4), and the hazard ratio (HR) of the three groups (named 0, 0.5 and 1, respectively) was calculated by univariate Cox analysis. Thus, HR represents the hazard ratio between groups 0–1–2/4 and 4/4 considering the intermediate groups such that 0–1–2/4 < 3/4 < 4/4. Patients with a score of 0–1–2/4, 3/4, and 4/4 had a median TFS of 107, 57 and 26 mo, respectively (HR = 4.03, p < 0.0001). These results were still significant in Binet stage A patients (HR = 4.64, p < 0.0001) (). For OS prediction, the multivariate stepwise Cox model showed SIRT5 and SIRT6 to be independent predictors (). We subsequently generated an OS score similar to the TFS score: when a patient was positive for SIRT5 or negative for SIRT6, one unit was added to the score. Surprisingly, HDAC3, SIRT2, SIRT3 and SIRT7, which were significant univariate predictors of OS, were not retained in this score, while SIRT5, which was not significant individually, was selected in multivariate analysis. Patients with an OS score of 0/2, 1/2 and 2/2 had a median OS of > 360, 237 and 94 mo, respectively (HR = 6.38, p < 0.0001) (). In Binet stage A, this score was close to the limit of significance but not verified, most likely because of the small number of events in this subgroup.
Table 1. Univariate and multivariate cox analysis for TFS and OS
Figure 3. TFS and OS score based on selected HDAC expression. TFS scores composed of HDAC6, 7 and 10 and SIRT3 selected by a multivariate stepwise Cox analysis were used to plot TFS with Kaplan-Meier curves for all Binet stages (A) and only Binet stage A (B). OS scores composed of SIRT5 and 6 selected by a multivariate stepwise Cox analysis were used to plot OS with Kaplan-Meier curves for all Binet stages (C) and only Binet stage A (D). The hazard ratio (HR) was calculated with univariate Cox regression.
Concerned by the risk of over fitting, we performed a 5-fold cross-validation study and observed a significant HR for the prediction of TFS (HR = 2.71, p = 0.0020) and OS (HR = 4.99, p = 0.0020), reinforcing our previous results (Fig. S3): the computation of this score was stable (concordance of 70.5% and 79.5% for TFS and OS, respectively). Additional details concerning cross-validation can be found in the Supplemental Material.
HDAC profile comparison between samples obtained at diagnosis and after relapse
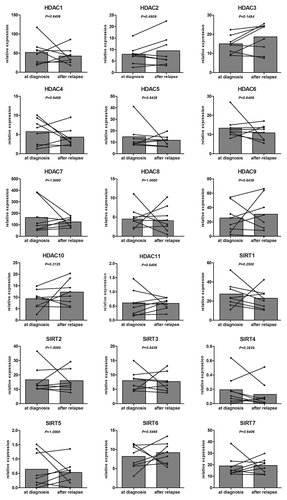
For eight patients, we compared samples obtained at diagnosis and after relapse in order to see if a relapse signature could be obtained. After performing a Wilcoxon non-parametric paired test, no significant differences could be highlighted (p > 0.05) ().
Figure 4. HDAC profile comparison between samples obtained at diagnosis and after relapse. Different HDAC isoenzyme expressions were plotted for 8 CLL patients for samples obtained at diagnosis and after relapse. HDAC values are expressed as relative fold change normalized to cyclophilin gene expression and calibrated with a common value. Significant differences were assessed using the Wilcoxon signed rank test.
Discussion
The aim of the present study was to evaluate the HDAC profile in CLL, particularly its clinical impact. First, CLL HDAC expression levels were compared with PB normal B cells. The normal counterpart of CLL cells is not clearly defined in the literature and remains controversial.Citation12 We therefore decided to add UCB samples in our experiments as a second control because they presented a higher proportion of CD5+CD19+ cells, which is similar to CLL cells. Other authors have used these cells as controls in CLL-based experiments.Citation13,Citation14 In addition, Gary-Gouy et al. have shown that CD5 molecule plays a role in the stimulation and survival of CLL cells,Citation15 and Saunders et al. observed that protein profiling classified CD5+ cord blood as the closest non-malignant counterpart of CLL.Citation16 Compared with both controls, HDAC-6, -7 and -11 and SIRT-3, -6 and -7 were statistically overexpressed in CLL samples. Overexpression of these genes has already been observed in other cancer types, such as breast cancer,Citation17,Citation18 oral squamous cell carcinoma,Citation19 pancreatic adenocarcinomaCitation20 and chronic myeloproliferative neoplasm,Citation21 suggesting that HDAC upregulation is often associated with cancer development. However, some HDACs (HDAC2 and SIRT4) were significantly downregulated in CLL compared with PB samples. Similar results have been observed for SIRT4 in acute myeloid leukemia,Citation22 but surprisingly, HDAC2 is generally upregulated in several cancer types.Citation6 These observations indicated that HDAC expression is highly variable from one cancer to another but also that their association with prognosis is not always found in all cancer types. However, in the present study, using combined analysis of HDAC, we were able to find robust and highly significant correlation with prognosis.
Only one report has investigated HDAC expression in CLL; however, our results are not in line with those obtained by Wang et al.Citation23 These important discrepancies can be explained by several parameters: the size of the CLL cohort (32 patients vs. 200 in our study), the purity of the samples (> 92% vs. a mean of 99%), the statistical test used (parametric student T test vs. non-parametric Mann-Whitney test) and the sensitivity of the quantitative real-time PCR (Ct mean values ranging from 34.3 to 39.2 vs. Ct mean values ranging from 25.8 to 35.2 in our study). The study of Wang et al. was most likely performed with too low cDNA amounts or with inefficient PCR because our samples had from 44- to 2,240-fold higher HDAC molecules per reaction (corresponding to 5.5 to 11.1 Ct difference compared with our study). In addition, Wang et al. assumed that all HDAC expressions had a Gaussian distribution, which is most likely not the case because a D'Agostino-Pearson normality test performed with our data (n = 200) indicated that HDAC expression did not fit to a Gaussian distribution (data not shown). We note the absence of data for HDAC4 and SIRT4 in that study. For all of these reasons, the results of Wang et al. are clearly debatable.
Because important variations exist between different HDAC levels (for example, high level of HDAC7, low level of SIRT4), HDAC expression levels were compared in different prognostic subgroups and correlated with TFS and OS data in our clinically well-characterized cohort. Surprisingly, low differences in expression were observed between poor and good prognosis patients according to several factors (ZAP70, CD38 and LPL), and a small number of these differences was statistically significant. Our results were again not in line with those of Wang et al.,Citation23 who observed differences between ZAP70+ and ZAP70- samples for HDAC-1, -3, -6, -7, -9, -11 and SIRT6. These discordances can again be explained by the small number of prognostic factor data (ZAP70, 20 cases vs. 200) and by the reasons reported above.
Correlation with TFS showed that a high level of HDAC6 was significantly associated with longer TFS, and a high level of HDAC3, SIRT2, SIRT3 and SIRT6 were associated with a longer OS. Overexpression of HDAC6 had also been associated with good prognosis in breast cancer,Citation17 cutaneous T-cell lymphomaCitation24 and lung cancer.Citation25 These observations are in accordance with those of Jung et al., who suggested that HDAC6 could be a tumor suppressor because ectopic overexpression of HDAC6 suppressed tumor cell growth and proliferation in various liver cancer cells.Citation26 In contrast, a high level of HDAC3 is often associated with poor prognosis in other cancers.Citation6 In the present study, SIRT2 and SIRT6 expression was also associated with good prognosis. To our knowledge, no study has associated the expression of SIRT6 with prognosis in any cancer, but interestingly, several lines of evidence suggest that SIRT6 acts as a guardian of genome stability, and it has therefore been proposed as a tumor suppressor.Citation27 Furthermore, Van Meter et al. demonstrate that overexpression of SIRT6 induces apoptosis in cancer cells but not in normal cells.Citation28 SIRT2 expression was already used in a 4-gene molecular prognostic signature in esophageal and junctional adenocarcinoma, and similar to our study, a high expression of SIRT2 was associated with good prognosis.Citation29 These results are consistent with the fact that SIRT2 can normally promote cell death, and its overexpression can also mediate a delay in cellular proliferation.Citation30 In addition, SIRT2 has been shown to suppress tumorigenesis by deacetylating co-activators of the anaphase-promoting complex/cyclosome activity.Citation31 SIRT3, meanwhile, has only been related in a study showing higher expression of this isoenzyme in node-positive breast cancer.Citation18 However, another team observed that SIRT3 protects against mitochondrial metabolism aberrations as reactive oxygen species (ROS) and therefore prevents genomic instability,Citation32 which could explain the prognostic advantage of this isoenzyme in our study. In summary, we showed here for the first time that high expression of HDAC-3, -6 and SIRT-2, -3 and -6 could have a positive effect on disease evolution. However, these results are counterintuitive since these HDACs are globally upregulated compared with one or both normal controls. Similar situations have already been observed in brain cancer where low HDAC6 is associated with high-grade glioma,Citation33 while global tumor sample display an HDAC upregulation compared with normal brain. The significance of these results is difficult to explain since the function, the target gene, the activity of HDAC could be different in malignant cells compared with normal cells. However, if we consider that, in general, low level of HDAC is associated with a higher level of global acetylation and consequently related to higher transcriptional activity, we could predict that gene transcription in CLL cells may be upregulated. In that case, we could infer that some oncogenes (such as anti-apoptotic genes) might be overexpressed and somehow leading to a poor prognosis. Functional explanations underlining these observations should be further investigated.
Because treatment and survival seem to be influenced by several HDACs, we combined HDAC-6, -7, -10 and SIRT3 (selected by a multivariate Cox stepwise analysis) in a TFS score and SIRT5 and 6 in an OS score. Interestingly, high levels of HDAC-7 and -10 and SIRT5 were associated with poor prognosis, whereas the opposite was observed for HDAC6 and SIRT-3 and -6. These observations are compatible with the work of Zhu et al. who showed that HDAC7 directly binds with c-Myc gene and that HDAC7 silencing decreased c-Myc mRNA highlighting the contribution of HDAC7 in cell proliferation and thus in tumor progression.Citation34 Moreover, HDAC10 have been shown to regulate Hsp90,Citation35 which stabilizes ZAP70, one of the most adverse prognosis factor in CLL patients.Citation36 These scores were able to clearly distinguish patients in three different subgroups, with median TFS ranging from 107 to 26 mo and median OS ranging from > 360 to 93 mo. These models were validated by a 5-fold cross-validation,Citation37 indicating that they were stable when the population was modified and that these results will most likely also be confirmed in another cohort. These data strongly suggested that concomitant or alternative HDAC expression could have an impact on CLL prognosis. This theory can be explained by the fact that different HDACs could target different promoter regions and could influence different gene expression. This situation is complicated by the fact that HDACs act in complex with other co-repressors/activators and can also acetylate crucial non-histone proteins (such as transcription factors like p53, p300, E2F and Nf-kBCitation38), thus regulating their activation and stability. Interestingly, it should be noted that even if an HDAC is expressed at low levels (such as SIRT5), it can still influence prognosis. Taken together, these data demonstrated why it is so difficult to explain the complex clinical significance of HDACs in CLL. Functional investigations of each HDAC individually are therefore needed.
The aim of this study was not to identify new prognostic factors but to highlight the role of epigenetic enzymes in the need for treatment, the survival of CLL patients and thus disease evolution. Functionally, HDAC inhibitors have already been shown to induce apoptosis of CLL cells in vitroCitation9,Citation10 and have been proposed as a new treatment strategy in CLL.Citation11 We can thus hypothesize that HDAC profiles could have an influence on treatment response and could potentially become a predictive marker. This question has not yet been addressed and should be validated in future clinical trials. When we compared samples of the same patients at diagnosis and after relapse, we were not able to find a signature linked to relapse. We can thus conclude that, even if some fluctuations could be observed, the emerging clone after disease relapse is not significantly different from diagnosis clone in terms of HDAC expression.
This report is the first comprehensive and complete quantitative study of HDAC expression linked to clinical data in CLL patients. Our results showed that HDAC expression is deregulated (mostly upregulated) in CLL patients compared with normal B cells. We also demonstrated that HDAC-3 and -6 and SIRT-2, -3 and -6 have an impact on TFS or OS when taken alone and that the specific combination of HDAC-6 and -7 and SIRT-7 and -10 for TFS and SIRT-5 and -6 for OS had complex prognostic significance. In conclusion, these data emphasize the important role of HDACs in CLL evolution and show that high and low expression of some HDACs could be associated with poor prognosis. These results could potentially be used in the future to predict the response of CLL patients to HDAC inhibitor treatment.
Materials and Methods
Patients and sample collection
This study was approved by the Bordet Institute Ethics Committee and conducted according to the principles expressed in the Declaration of Helsinki. All of the samples were collected after obtaining written informed consent from 200 CLL patients who presented with a typical CD19+CD5+CD23+ phenotype. The median age of this population was 63 y (range 34–89). All of the samples were collected at the time of diagnosis before any treatment. For eight patients, we also compared HDAC profiles on samples obtained at diagnosis and after relapse. Control samples were obtained from peripheral blood (PB) of 20 age-matched healthy volunteers (median age of 64 y; range: 46–90) after written informed consent was obtained and from 20 umbilical cord blood (UCB) samples after full-term delivery and after obtaining written informed consent of the mothers. A summary of the patient characteristics is presented in Table S4. Treatment-free survival (TFS) and overall survival (OS) were calculated from the time of diagnosis until the date of first treatment and the date of death, respectively. All of the deaths were CLL-related. Of the 98 (100%) patients who received a treatment, 36 (37%) patients received alkylating agent-based treatment (chlorambucil alone, mini-CHOP,…), 20 (20%) patients received fludarabine-based (without rituximab) treatment (fludarabine alone, fludarabine-cyclophosphamide,…), 28 (29%) patients received FCR and finally 14 (14%) patients received other treatment types (lenalidomide, lumiliximab, campath,…). However no statistical impact of these treatments was observed on OS (data not shown). The median TFS of this cohort was 85 mo (range, 0.03–244), whereas the median OS was 242 mo (range, 0.4–360). The median follow-up was 77 mo (range 0.4–360).
RNA extraction, cDNA synthesis and quantification of HDAC isoenzymes
Peripheral blood mononuclear cells were isolated by density gradient centrifugation with Linfosep (Biomedics). B cells were purified with a CD19+ magnetic bead system (MidiMACS, Miltenyi Biotec) according to the manufacturer's instructions. The mean B cell purity for the 200 CLL samples was 99.0% (range 94.2–100), 97.4% for the 20 PB samples (range 95.7–99.0) and 98.6% for the 20 UCB samples (range 96.4–99.5), as measured by flow cytometry. The mean percentage of CD19+CD5+ cells in CLL samples was 96.8% (range 70.0–100), 6.8% in PB samples (range 0.7–12.8), and 56.3% in UCB samples (range 21.4–90.8). Total RNA was extracted from purified CD19+ cells in a single step using TriPure Isolation Reagent (Roche Applied Science). cDNA was generated from 500 ng of RNA using qScript cDNA supermix (Quanta Biosciences) according to the manufacturer’s protocol.
The HDAC expression profile was quantified by real-time PCR using custom Taqman® low density arrays (TLDA, Life Technologies-Applied Biosystems): the “format 48” chosen for this 384-well card included 8 lines of wells pre-coated (in duplicate) with specific primers and a TaqMan probe for the 18 HDACs and the cyclophilin (PPIA) gene as an endogenous control. Each line was loaded with 100 µl containing 100 ng of cDNA in 50 µl of water and 50 µl of TaqMan® Universal PCR Master Mix (Applied Biosystems). The card was centrifuged 2 times at 1,200 rpm for 1 min and sealed. Standard real-time PCR was performed with a ViiA™ 7 Real-Time PCR System (Applied Biosystems). To ensure that all expression levels were comparable, HDAC expression was normalized with the cyclophilin A gene as an endogenous control and calibrated by subtracting 10 (chosen arbitrarily) from the ΔCt. The comparative ΔΔCt method was then applied for data analysis, and fold changes were subsequently calculated (fold change = 2-ΔΔCt).
Assessment of other prognostic factors
ZAP70 and LPL were measured by real-time PCR as previously described.Citation39 CD38 expression was assessed by flow cytometry, sCD23 and β2-M were assessed by ELISA, and IgVH gene mutational analysis was performed as previously described.Citation40 LDT was assessed according to Montserrat et al.Citation41 Classical cytogenetics by standard karyotype analysis and additional interphase FISH were performed to screen for the most common aberrations using Chromoprobe Multiprobe® - CLL System (Cytocell, Amplitech). Patients were then classified according to Döhner's recommendations.Citation42 Additional details can be found in the Supplemental Materials. All of these factors were proven to be significant predictors of TFS and OS, indicating that our cohort is representative of a CLL population (Fig. S4 and Table S4)
Statistical analysis
We performed ROC curves to determine the cut-off values of each HDAC that best distinguished ZAP70+ and ZAP70- cases and minimized the number of false negatives. ZAP70 was used in ROC curve analysis to maximize the concordance with prognosis because ZAP70 has been shown as one of the most powerful prognostic factors in our previous studies.Citation39,Citation40,Citation43 Significant differences between groups were assessed using the Mann-Whitney non-parametric test or with Wilcoxon signed rank test for paired experiments. TFS and OS distributions were plotted using Kaplan-Meier estimates and compared using the log-rank test. Univariate Cox regression analysis evaluated the effects of the different prognostic variables on TFS or OS. Multivariate Cox regression stepwise analysis was performed using all binarized HDAC values to investigate the possible concomitant role of HDAC expression in CLL prognosis and to generate a score for TFS and OS prediction. Because a validation set was not available, a 5-fold cross-validation study was performed to assess overfitting risk and the stability of the HDAC scoreCitation37 (see Supplemental Materials for details). All of the tests were two-sided. An effect was considered to be statistically significant at p < 0.05. All of the analyses were performed with GraphPad Prism 5.0 (Graph-Pad Software) or SPSS 15.0 software.
Additional material
Download Zip (652.6 KB)Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Financial Disclosures
This research was supported by the “Fonds IRIS-Recherche” and the “Télévie Fund.” B.S. (Postdoctoral Researcher) and L.L. (Senior Research Associate) are members of the F.N.R.S.
Author Contributions
M.V.D. and B.S. performed the research and statistical analysis, analyzed the data, made the figures and tables, and wrote the manuscript. E.C. participated to the revision of the manuscript. N.M., P.M. and D.B. helped provide the patient samples and data. B.S. and L.L. performed, supervised and designed the research and revised the manuscript.
Supplemental Materials
Supplemental materials may be found here: www.landesbioscience.com/journals/epigenetics/article/22674
References
- Workman JL, Kingston RE. Alteration of nucleosome structure as a mechanism of transcriptional regulation. Annu Rev Biochem 1998; 67:545 - 79; http://dx.doi.org/10.1146/annurev.biochem.67.1.545; PMID: 9759497
- Forsberg EC, Bresnick EH. Histone acetylation beyond promoters: long-range acetylation patterns in the chromatin world. Bioessays 2001; 23:820 - 30; http://dx.doi.org/10.1002/bies.1117; PMID: 11536294
- Minucci S, Pelicci PG. Histone deacetylase inhibitors and the promise of epigenetic (and more) treatments for cancer. Nat Rev Cancer 2006; 6:38 - 51; http://dx.doi.org/10.1038/nrc1779; PMID: 16397526
- Witt O, Deubzer HE, Milde T, Oehme I. HDAC family: What are the cancer relevant targets?. Cancer Lett 2009; 277:8 - 21; http://dx.doi.org/10.1016/j.canlet.2008.08.016; PMID: 18824292
- Secrist JP, Zhou X, Richon VM. HDAC inhibitors for the treatment of cancer. Curr Opin Investig Drugs 2003; 4:1422 - 7; PMID: 14763127
- Weichert W. HDAC expression and clinical prognosis in human malignancies. Cancer Lett 2009; 280:168 - 76; http://dx.doi.org/10.1016/j.canlet.2008.10.047; PMID: 19103471
- Hamblin T. Chronic lymphocytic leukaemia: one disease or two?. Ann Hematol 2002; 81:299 - 303; http://dx.doi.org/10.1007/s00277-002-0476-1; PMID: 12107557
- Van Bockstaele F, Verhasselt B, Philippé J. Prognostic markers in chronic lymphocytic leukemia: a comprehensive review. Blood Rev 2009; 23:25 - 47; http://dx.doi.org/10.1016/j.blre.2008.05.003; PMID: 18599169
- Stamatopoulos B, Meuleman N, De Bruyn C, Mineur P, Martiat P, Bron D, et al. Antileukemic activity of valproic acid in chronic lymphocytic leukemia B cells defined by microarray analysis. Leukemia 2009; 23:2281 - 9; http://dx.doi.org/10.1038/leu.2009.176; PMID: 19710697
- Stamatopoulos B, Meuleman N, De Bruyn C, Delforge A, Bron D, Lagneaux L. The histone deacetylase inhibitor suberoylanilide hydroxamic acid induces apoptosis, down-regulates the CXCR4 chemokine receptor and impairs migration of chronic lymphocytic leukemia cells. Haematologica 2010; 95:1136 - 43; http://dx.doi.org/10.3324/haematol.2009.013847; PMID: 20145270
- Stimson L, Wood V, Khan O, Fotheringham S, La Thangue NB. HDAC inhibitor-based therapies and haematological malignancy. Ann Oncol 2009; 20:1293 - 302; http://dx.doi.org/10.1093/annonc/mdn792; PMID: 19515748
- Caligaris-Cappio F, Ghia P. The normal counterpart to the chronic lymphocytic leukemia B cell. Best Pract Res Clin Haematol 2007; 20:385 - 97; http://dx.doi.org/10.1016/j.beha.2007.02.005; PMID: 17707828
- Farren TW, Giustiniani J, Liu FT, Tsitsikas DA, Macey MG, Cavenagh JD, et al. Differential and tumor-specific expression of CD160 in B-cell malignancies. Blood 2011; 118:2174 - 83; http://dx.doi.org/10.1182/blood-2011-02-334326; PMID: 21715317
- Gutierrez AJ Jr., Tschumper RC, Wu X, Shanafelt TD, Eckel-Passow J, Huddleston PM 3rd, et al. LEF-1 is a prosurvival factor in chronic lymphocytic leukemia and is expressed in the preleukemic state of monoclonal B-cell lymphocytosis. Blood 2010; 116:2975 - 83; http://dx.doi.org/10.1182/blood-2010-02-269878; PMID: 20595513
- Gary-Gouy H, Sainz-Perez A, Marteau JB, Marfaing-Koka A, Delic J, Merle-Beral H, et al. Natural phosphorylation of CD5 in chronic lymphocytic leukemia B cells and analysis of CD5-regulated genes in a B cell line suggest a role for CD5 in malignant phenotype. J Immunol 2007; 179:4335 - 44; PMID: 17878328
- Saunders FK, Lawry J, Winfield DA, Goepel JR, Hancock BW, Sharrard RM, et al. Comparison of protein synthesis profiles in chronic lymphocytic leukaemia cells and B-lymphocytes from peripheral blood, cord blood and tonsil. Experientia 1994; 50:493 - 6; http://dx.doi.org/10.1007/BF01920755; PMID: 7515011
- Zhang Z, Yamashita H, Toyama T, Sugiura H, Omoto Y, Ando Y, et al. HDAC6 expression is correlated with better survival in breast cancer. Clin Cancer Res 2004; 10:6962 - 8; http://dx.doi.org/10.1158/1078-0432.CCR-04-0455; PMID: 15501975
- Ashraf N, Zino S, Macintyre A, Kingsmore D, Payne AP, George WD, et al. Altered sirtuin expression is associated with node-positive breast cancer. Br J Cancer 2006; 95:1056 - 61; http://dx.doi.org/10.1038/sj.bjc.6603384; PMID: 17003781
- Sakuma T, Uzawa K, Onda T, Shiiba M, Yokoe H, Shibahara T, et al. Aberrant expression of histone deacetylase 6 in oral squamous cell carcinoma. Int J Oncol 2006; 29:117 - 24; PMID: 16773191
- Ouaïssi M, Sielezneff I, Silvestre R, Sastre B, Bernard JP, Lafontaine JS, et al. High histone deacetylase 7 (HDAC7) expression is significantly associated with adenocarcinomas of the pancreas. Ann Surg Oncol 2008; 15:2318 - 28; http://dx.doi.org/10.1245/s10434-008-9940-z; PMID: 18506539
- Skov V, Larsen TS, Thomassen M, Riley CH, Jensen MK, Bjerrum OW, et al. Increased gene expression of histone deacetylases in patients with Philadelphia-negative chronic myeloproliferative neoplasms. Leuk Lymphoma 2012; 53:123 - 9; http://dx.doi.org/10.3109/10428194.2011.597905; PMID: 21806350
- Bradbury CA, Khanim FL, Hayden R, Bunce CM, White DA, Drayson MT, et al. Histone deacetylases in acute myeloid leukaemia show a distinctive pattern of expression that changes selectively in response to deacetylase inhibitors. Leukemia 2005; 19:1751 - 9; http://dx.doi.org/10.1038/sj.leu.2403910; PMID: 16121216
- Wang JC, Kafeel MI, Avezbakiyev B, Chen C, Sun Y, Rathnasabapathy C, et al. Histone deacetylase in chronic lymphocytic leukemia. Oncology 2011; 81:325 - 9; http://dx.doi.org/10.1159/000334577; PMID: 22237050
- Marquard L, Gjerdrum LM, Christensen IJ, Jensen PB, Sehested M, Ralfkiaer E. Prognostic significance of the therapeutic targets histone deacetylase 1, 2, 6 and acetylated histone H4 in cutaneous T-cell lymphoma. Histopathology 2008; 53:267 - 77; http://dx.doi.org/10.1111/j.1365-2559.2008.03109.x; PMID: 18671804
- Osada H, Tatematsu Y, Saito H, Yatabe Y, Mitsudomi T, Takahashi T. Reduced expression of class II histone deacetylase genes is associated with poor prognosis in lung cancer patients. Int J Cancer 2004; 112:26 - 32; http://dx.doi.org/10.1002/ijc.20395; PMID: 15305372
- Jung KH, Noh JH, Kim JD, Eun JW, Bae JH, Chang Y, et al. HDAC6 functions as a tumor suppressor by activating JNK-mediated beclin 1-dependent autophagic cell death in liver cancer. Hepatology 2012; 56:644 - 57; http://dx.doi.org/10.1002/hep.25699; PMID: 22392728
- Bosch-Presegué L, Vaquero A. The dual role of sirtuins in cancer. Genes Cancer 2011; 2:648 - 62; http://dx.doi.org/10.1177/1947601911417862; PMID: 21941620
- Van Meter M, Mao Z, Gorbunova V, Seluanov A. SIRT6 overexpression induces massive apoptosis in cancer cells but not in normal cells. Cell Cycle 2011; 10:3153 - 8; http://dx.doi.org/10.4161/cc.10.18.17435; PMID: 21900744
- Peters CJ, Rees JRE, Hardwick RH, Hardwick JS, Vowler SL, Ong CA, et al, Oesophageal Cancer Clinical and Molecular Stratification (OCCAMS) Study Group. A 4-gene signature predicts survival of patients with resected adenocarcinoma of the esophagus, junction, and gastric cardia. Gastroenterology 2010; 139:1995 - 2004, e15; http://dx.doi.org/10.1053/j.gastro.2010.05.080; PMID: 20621683
- Inoue T, Hiratsuka M, Osaki M, Oshimura M. The molecular biology of mammalian SIRT proteins: SIRT2 in cell cycle regulation. Cell Cycle 2007; 6:1011 - 8; http://dx.doi.org/10.4161/cc.6.9.4219; PMID: 17457050
- Kim HS, Vassilopoulos A, Wang RH, Lahusen T, Xiao Z, Xu X, et al. SIRT2 maintains genome integrity and suppresses tumorigenesis through regulating APC/C activity. Cancer Cell 2011; 20:487 - 99; http://dx.doi.org/10.1016/j.ccr.2011.09.004; PMID: 22014574
- Park SH, Ozden O, Jiang H, Cha YI, Pennington JD, Aykin-Burns N, et al. Sirt3, Mitochondrial ROS, Ageing, and Carcinogenesis. Int J Mol Sci 2011; 12:6226 - 39; http://dx.doi.org/10.3390/ijms12096226; PMID: 22016654
- Lucio-Eterovic AKB, Cortez MAA, Valera ET, Motta FJN, Queiroz RGP, Machado HR, et al. Differential expression of 12 histone deacetylase (HDAC) genes in astrocytomas and normal brain tissue: class II and IV are hypoexpressed in glioblastomas. BMC Cancer 2008; 8:243; http://dx.doi.org/10.1186/1471-2407-8-243; PMID: 18713462
- Zhu C, Chen Q, Xie Z, Ai J, Tong L, Ding J, et al. The role of histone deacetylase 7 (HDAC7) in cancer cell proliferation: regulation on c-Myc. J Mol Med (Berl) 2011; 89:279 - 89; http://dx.doi.org/10.1007/s00109-010-0701-7; PMID: 21120446
- Park JH, Kim SH, Choi MC, Lee J, Oh DY, Im SA, et al. Class II histone deacetylases play pivotal roles in heat shock protein 90-mediated proteasomal degradation of vascular endothelial growth factor receptors. Biochem Biophys Res Commun 2008; 368:318 - 22; http://dx.doi.org/10.1016/j.bbrc.2008.01.056; PMID: 18211808
- Castro JE, Prada CE, Loria O, Kamal A, Chen L, Burrows FJ, et al. ZAP-70 is a novel conditional heat shock protein 90 (Hsp90) client: inhibition of Hsp90 leads to ZAP-70 degradation, apoptosis, and impaired signaling in chronic lymphocytic leukemia. Blood 2005; 106:2506 - 12; http://dx.doi.org/10.1182/blood-2005-03-1099; PMID: 15972449
- Höfling H, Tibshirani R. A study of pre-validation. Ann.Appl.Stat. 2008; 2:643 - 64; http://dx.doi.org/10.1214/07-AOAS152
- Ropero S, Esteller M. The role of histone deacetylases (HDACs) in human cancer. Mol Oncol 2007; 1:19 - 25; http://dx.doi.org/10.1016/j.molonc.2007.01.001; PMID: 19383284
- Stamatopoulos B, Meuleman N, Haibe-Kains B, Duvillier H, Massy M, Martiat P, et al. Quantification of ZAP70 mRNA in B cells by real-time PCR is a powerful prognostic factor in chronic lymphocytic leukemia. Clin Chem 2007; 53:1757 - 66; http://dx.doi.org/10.1373/clinchem.2007.089326; PMID: 17702857
- Stamatopoulos B, Meuleman N, De Bruyn C, Pieters K, Anthoine G, Mineur P, et al. A molecular score by quantitative PCR as a new prognostic tool at diagnosis for chronic lymphocytic leukemia patients. PLoS One 2010; 5:5; http://dx.doi.org/10.1371/journal.pone.0012780; PMID: 20862275
- Montserrat E, Sanchez-Bisono J, Viñolas N, Rozman C. Lymphocyte doubling time in chronic lymphocytic leukaemia: analysis of its prognostic significance. Br J Haematol 1986; 62:567 - 75; http://dx.doi.org/10.1111/j.1365-2141.1986.tb02969.x; PMID: 3954968
- Döhner H, Stilgenbauer S, Benner A, Leupolt E, Kröber A, Bullinger L, et al. Genomic aberrations and survival in chronic lymphocytic leukemia. N Engl J Med 2000; 343:1910 - 6; http://dx.doi.org/10.1056/NEJM200012283432602; PMID: 11136261
- Stamatopoulos B, Meuleman N, Haibe-Kains B, Saussoy P, Van Den Neste E, Michaux L, et al. microRNA-29c and microRNA-223 down-regulation has in vivo significance in chronic lymphocytic leukemia and improves disease risk stratification. Blood 2009; 113:5237 - 45; http://dx.doi.org/10.1182/blood-2008-11-189407; PMID: 19144983