Abstract
Keap1 (Kelch-like ECH-associated protein 1) is an adaptor protein that mediates the ubiquitination/degradation of genes regulating cell survival and apoptosis under oxidative stress conditions. We determined methylation status of the KEAP1 promoter in 102 primary breast cancers, 14 pre-invasive lesions, 38 paired normal breast tissues and 6 normal breast from reductive mammoplasty by quantitative methylation specific PCR (QMSP). Aberrant promoter methylation was detected in 52 out of the 102 primary breast cancer cases (51%) and 10 out of 14 pre-invasive lesions (71%). No mutations of the KEAP1 gene were identified in the 20 breast cancer cases analyzed by fluorescence based direct sequencing. Methylation was more frequent in the subgroup of patients identified as ER positive-HER2 negative tumors (66.7%) as compared with triple-negative breast cancers (35%) (p = 0.05, Chi-square test). The impact of the interactions between Er, PgR, Her2 expression and KEAP1 methylation on mortality was investigated by RECPAM multivariable statistical analysis, identifying four prognostic classes at different mortality risks. Triple-negative breast cancer patients with KEAP1 methylation had higher mortality risk than patients without triple-negative breast cancer (HR = 14.73, 95%CI: 3.65–59.37). Both univariable and multivariable COX regressions analyses showed that KEAP1 methylation was associated with a better progression free survival in patients treated with epirubicin/cyclophosfamide and docetaxel as sequential chemotherapy (HR = 0.082; 95%CI: 0.007–0.934). These results indicate that aberrant promoter methylation of the KEAP1 gene is involved in breast cancerogenesis. In addition, identifying patients with KEAP1 epigenetic abnormalities may contribute to disease progression prediction in breast cancer patients.
Introduction
The Keap1 (Kelch-like ECH-associated protein 1) protein is emerging as a main negative regulator of the cell adaptive response to oxidative stress. Keap1 possesses a dual role because it is able to sense a disturbance in the redox homeostasis and switch on and off its well-known substrate, Nrf2 (nuclear factor-erythroid 2-related factor 2) transcription factor.Citation1-Citation4 Under physiological conditions, Nrf2 is retained in the cytoplasm by the binding with Keap1 and it is maintained at a reduced level by the Keap1-dependent ubiquitination and proteasomal degradation systems. Under exposure to oxidative stress, Keap1-dependent ubiquitin ligase activity is inhibited and Nrf2 can translocate into the nucleus, where it binds to antioxidant response elements (ARE) located in the promoter of detoxifying genes.Citation5 The first link between Keap1 and cancerogenesis was the identification of somatic gene mutations leading to a permanent Nrf2 activation in non-small cell lung cancer (NSCLC).Citation6-Citation8 Indeed, recent studies suggest that Keap1 may regulate several cellular pathways deeply involved in carcinogenesis. One of the Keap1 substrates for ubiquitination is Ikkβ which upregulates the Nf-κB transcription factor through the negative regulation of its inhibitor Ikbα.Citation9 Under oxidative stress, Ikkβ is released from Keap1, and mediates the activation of Nf-kB regulated genes, resulting in increased growth, proliferation and anti-apoptosis, thus contributing to cell survival and tumor progression.Citation10 Moreover, recent studies suggest that under oxidative stress conditions Keap1 modulates the intrinsic apoptotic pathway through the degradation of Bcl2 and Bcl-xL anti-apoptotic proteins. While Keap1 directly binds Bcl2, the effect on Bcl-xL is mediated by the interaction with Pgam5 protein, which acts as a bridge between Keap1 and Bcl-xL.Citation11,Citation12 Overall, these data suggest that Keap1 may act as a tumor suppressor gene and loss of Keap1 functions confers tumorigenic potential to the cells.
The frequency of KEAP1 somatic mutations in the Nrf2 interacting domain is extensively variable among the tumor types analyzed. The highest frequency of mutations was found in gallbladder cancer (30.7%), ovarian clear cell carcinoma (29%) and NSCLC (19%).Citation6-Citation8,Citation13-Citation20 Lower frequencies of mutations were demonstrated in endometrioid endometrial tumors (8.5%), non-clear cell ovarian cancer (8%), hepatocellular carcinoma (8.9%), colorectal cancer (7.8%), biliary duct carcinomas (5%), breast cancer (2%), prostate cancer (1.3%) and gastric cancer (1%).Citation13,Citation17,Citation21,Citation22 However, immunohistochemical studies in lung cancer, endometrial tumors, renal and breast cancer demonstrated a high frequency of KEAP1 downregulation and/or Nrf2 overexpression in these tumor types, suggesting that the deregulation of KEAP1 may play a role in carcinogenesis beside the presence of genomic alterations.Citation19,Citation23-Citation25 We have recently confirmed that epigenetic modification by promoter methylation is a main mechanism of regulation of KEAP1 gene expression. Aberrant methylation of the KEAP1 promoter was found in approximately 50% of NSCLC and in 70% of malignant gliomas.Citation18,Citation26 The presence of two genetic or epigenetic abnormalities was associated with worst progression free survival in patients affected by NSCLC,Citation18 whereas concomitant methylation of MGMT and KEAP1 genes was able to better predict response to treatment with radiotherapy and temozolomide in malignant gliomas.Citation26 Hanada et al.Citation27 recently reported aberrant promoter methylation in 53% of colorectal cancer without finding correlation with patients’ survival.
In the present study we evaluated the frequency of KEAP1 genetic and epigenetic changes in a series of primary breast tumors and pre-invasive breast lesions. We found aberrant promoter methylation in 51% of the cases without finding somatic gene mutations in the subset of tumors analyzed. Aberrant promoter methylation was associated with a higher risk of dying from the disease in triple-negative breast cancer. When we correlated KEAP1 methylation with different chemotherapy schemes, it was a predictor of lower risk of tumor relapse in the group of patients treated with epirubicin/cyclophosfamide followed by docetaxel.
Results
Patients and treatment
Table S1 summarizes descriptive statistics of the 102 cases selected for the analysis. The median age of the study population was 57 y (range 31 to 85 y) and the median tumor size was 2.2 cm (range 1.0 to 8.0 cm). Metastases at diagnosis were present in 6 cases whereas, among non-metastatic patients, 23 experienced disease progression within the follow up time. Of those, 22 developed distant metastases and 1 local recurrence. First metastatic sites were bone (n = 16), lung (n = 6), brain (n = 3) and liver (n = 3). Of the 28 patients with synchronous (n = 6) or metachronous (n = 22) distant metastases, 17 died and 11 are currently alive with disease. Median follow up data for living patients is 56 mo (IQR: 40–65 mo).
All patients received adequate local treatment (breast conserving surgery or total mastectomy) plus sentinel node biopsy or complete axillary dissection. Post-surgery treatments were performed according to the following guidelines: San Gallen, NCCN and ASCO. Postoperative breast irradiation (RT) was performed in 64 out of the 102 patients. All 96 patients without metastases at diagnosis received adjuvant treatment: 17 hormone therapy alone, 45 hormone therapy in combination with chemotherapy and 34 chemotherapy alone. Of the 62 patients receiving hormone therapy, 25 were treated with tamoxifen (40.3%), 30 with aromatase inhibitors (48.4%) and 7 started treatment with tamoxifen and then switched on aromatase inhibitors (11.3%). All metastatic patients received at least one first line chemotherapy treatment. Of the 16 patients bearing or developing bone metastases, 6 received zolendronate. Anti-Her2 therapy with monoclonal antibody was performed in 14 patients as adjuvant treatment.
Aberrant methylation of KEAP1 promoter is a frequent and early event in breast cancer
We determined KEAP1 methylation levels in normal tissues and tumor specimens by using a primers/probe set designed to amplify a CpG region showing the highest frequency of methylation and correlating with KEAP1 mRNA expression.Citation18,Citation26,Citation28 Methylation analysis was performed on DNA obtained from 6 normal breast tissues from reductive mammoplasty (NB), 14 pre-invasive lesions (PreINV), 102 invasive tumors (INV) along with 38 paired normal breast tissues distant from tumor (NBDT). Of the pre-invasive lesions, 12 were synchronous (9 DCIS and 3 ADH) to invasive cancer and 2 were only DCIS. The median values and InterQuartile Ranges (IQR) of KEAP1/ACTB ratios were 0 (IQR: 0–0.07) for normal breast tissues (NB), 0 (IQR: 0–0) for normal breast tissues distant from tumors (NBDT), 1.68 (IQR: 0.46–10.07) for pre-invasive lesions (PreINV) and 0.23 (IQR 0–7.46) for invasive tumors (INV) (). No statistically significant differences in methylation levels resulted between NBDT and NB groups. However, statistically significant differences were found when NBDT, PreINV and INV groups were compared (p < 0.001, Skillings and Mack test). The analysis between groups showed significantly higher methylation levels in the invasive (INV) breast lesions as compared with NBDT (post-hoc pairwise comparison p < 0.001). Nevertheless, no significant differences were found between PreINV and NBDT as well as between PreINV and INV groups.
Figure 1. Boxplots of KEAP1 promoter methylation levels in normal breast tissues from reductive mammoplasty (NB), normal breast distant from tumor (NBDT), pre invasive lesions (PreINV) and invasive cancer (INV). Methylation levels are expressed as the KEAP1/ACTB ratio multiplied by 1,000. The boxes mark the interquartile range, (interval between the 25th and 75th percentile). The lines inside the boxes denote median values, *denote the outliers.
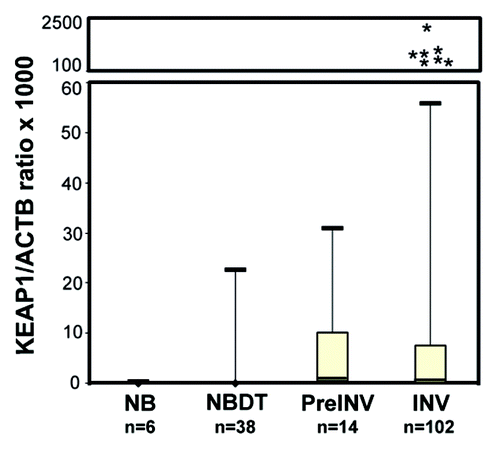
The discriminatory power of the KEAP1 QMSP assay was assessed by estimating the area under the ROC curve. Paired normal breast tissues distant from tumor (NBDT) and invasive lesions (INV) were used to estimate the ROC curve. The AUC value was 0.686 (95%CI: 0.620–0.751) (Fig. S1) with an optimal cut off value of 0.22, a sensitivity of 51% (95%CI: 41.4–0.60%) and a specificity of 89.47% (95%CI: 76–96%). In pre-invasive lesions, methylation was detected in 10 out of 14 cases (71.4%), whereas 1 of the 6 normal breast tissues from reductive mammoplasty was methylated (17%). In all cases the same methylation status was demonstrated in pathological specimens and paired normal breast. Only one of the cases for which pre-invasive and invasive lesions were both available showed methylation in the tumor but not in the paired DCIS.
To evaluate the possible involvement of genetic abnormalities in KEAP1 deregulation in breast cancer, we analyzed 20 cases by sequencing the portion of KEAP1 gene encoding for the Double Glycin Region (DGR) domain, which contains binding sites for Nrf2, Bcl2, Pgam5 and Ikkβ. This analysis did not reveal any sequence variations that would likely result in a functional alteration in Keap1 protein.
KEAP1 methylation is associated with high risk of death in triple-negative breast cancers (TNBC)
As shown in , we evaluated whether KEAP1 methylation status defined by the cut off value was associated with patients’ clinicopathological characteristics. The only statistically significant association was found between KEAP1 methylation status and breast cancer classification based on receptors status (p = 0.05) (). In particular, aberrant methylation was more frequent in the subgroup of patients identified as Er-positive/HER2-negative tumors (66.7%) and less frequent in the subgroup of triple-negative breast cancers (TBNC, 35%) (p = 0.05, Chi square test).
Table 1. Associations between KEAP1 methylation above cut off value and patients clinical pathological characteristics (n = 102)
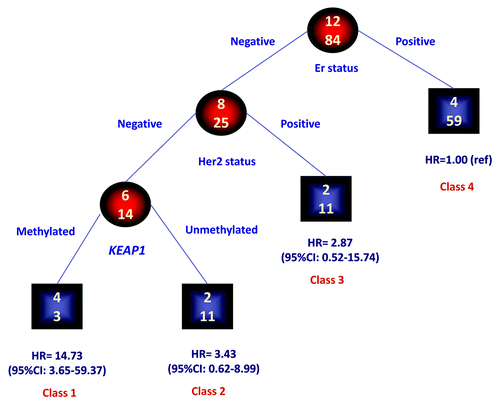
We evaluated, in the group of patients without metastases at diagnosis (n = 96), whether KEAP1 methylation adds prognostic information to the breast cancer classification applying the RECPAM method. The tree-growing algorithm evaluates the interactions between KEAP1 methylation and Er, PgR and HER2 status and modeled hazard ratios from a COX regression analysis. As shown in , four prognostic classes of breast cancer at different risks of death for the disease were identified. The reference class is represented by the subgroup with the lowest incidence of death events. Thus, the hazard ratio (HR) for all the other subgroups was estimated with respect to the reference class. The most important variable in discriminating the risk of death was Er status (Class 4), whereas PgR status was not able to discriminate prognostic classes. Er-negative/HER2-positive patients (Class 3) showed a better prognosis when compared with Er-negative/HER2-negative patients. In the latter group, the risk of death was further discriminated by KEAP1 status. Indeed methylated KEAP1 patients showed the highest risk of cancer related deaths (Class 1, HR = 14.73, 95%CI: 3.65–59.37). It is of note that this category is also identified as triple-negative breast cancers (TNBC), because in our cohort none of the cases showed an Er-negative/PgR-positive phenotype.
Figure 2. Identification of subgroups at different risks for mortality: results of RECPAM analysis. RECPAM analysis identified patient subgroups at different risks for mortality. The tree-growing algorithm modeled hazard ratios after a Cox proportional hazards regression model with KEAP1, ER, PgR and HER2 status as candidate splitting variables. Chosen splitting variables are shown between branches, while condition sending patients to left or right sibling is on relative branch. Class 4 with the lowest mortality risk was reference category (HR = 1). Circles indicate subgroups of patients. Squares indicate patient subgroup RECPAM class. Numbers inside circles and squares represent the number of events (top) and the number of non-events (bottom), respectively.
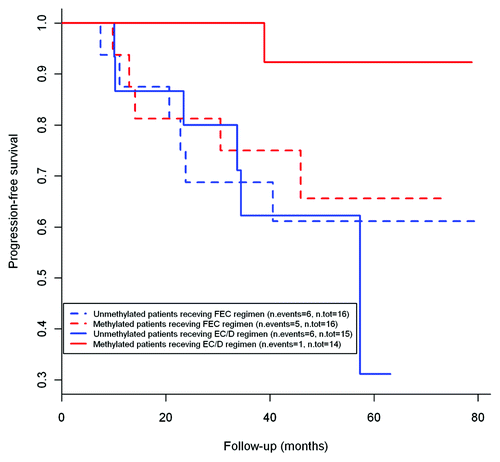
KEAP1 methylation is associated with better progression free survival (PFS) in patients treated with epirubicin/cyclofosfamide and docetaxel regimen
We evaluated whether KEAP1 methylation status may be predictive of outcome in patients receiving different schemes of treatment. A difference in progression free survival (PFS) was found among KEAP1 methylated and unmethylated patients receiving sequential therapy with epirubicin/cyclophoshamide followed by taxanes (EC/D) (n = 30). Univariable COX regression analysis showed an association between KEAP1 methylation and better PFS (HR = 0.096; 95%CI: 0.011–0.832; p = 0.03) This result was confirmed in multivariable analysis using lymph node status, HER2 status and Ki67/Mib1 as adjustment variables (HR = 0.082; 95%CI: 0.007–0.934, p = 0.04). As shown in , Kaplan-Meier analysis for PFS indicate that KEAP1 methylated patients treated with EC/D have the better prognosis as compared with unmethylated EC/D treated subjects and patients receiving 5-fluorouracyl, epirubicyn and cyclophosfamide (FEC, n = 32). Overall survival (OS) estimates could not be computed due to the low number of events in the population study (n = 3). The analysis could not be performed, due to the low number of cases, neither in patients treated with cyclophosfamide, metotrexate and 5-fluorouracyl (CMF n = 11) nor in the group of patients treated with Epirubicin/vinorelbin (EPI/VNR)as neoadjuvant therapy (n = 6).
Discussion
We investigated the possible contribution of KEAP1 genetic and epigenetic abnormalities to development and progression of breast cancer. Aberrant promoter methylation was detected in approximately half of the samples by QMSP analysis. Since a previous studyCitation17 identified a low frequency of somatic mutations in the KEAP1 gene in breast tumors, we sequenced the gene coding regions corresponding to Nrf2, Bcl2, Pgam5 and Ikkβ protein binding sites in 20 breast cancer cases without finding any pathological variation, further suggesting that KEAP1 somatic mutations are an infrequent event in this tumor type. Eleven of the 12 cases for which paired pre-invasive lesions (ADH and DCIS) were available showed the same methylation status of synchronous primary tumor, suggesting that KEAP1 promoter methylation is an early event in breast carcinogenesis. We compared methylation status with tumor pathological characteristics finding that methylation was less frequent in triple-negative breast cancer (TNBC) as compared with Er-positive/HER2-negative tumors. This result is consistent with a previous report in which TNBC showed higher frequency of Keap1 protein overexpression as compared with Er-positive/HER2-negative tumors.Citation24 In addition, in our cohort of TNBC patients, KEAP1 methylation was associated with an increased risk of cancer related death by using a multivariable model including hormone receptor status, HER2, Ki67. Estrogens are key carcinogenic factors for the development of breast cancer and one of the non-genomic mechanisms for estrogen carcinogenesis lies in the oxidation to semiquinones and quinones by cytochrome 450 enzymes.Citation29,Citation30 The induction of the Nrf2 detoxifying enzyme NAD(P)H oxidoreductase (NQO1) is an important mechanism of protection from estrogen-induced damage. Anti-estrogens agents stimulate NQO1 transcription and protect against estrogen-induced DNA damage in estrogen-dependent breast cancer cells.Citation31,Citation32 It has been observed that the Erα binding domain is required for estrogens inhibition of the NQO1 promoter activity leading to repression of its transcription.Citation33 Thus, in Er-positive tumors the upregulation of Nrf2 by increased KEAP1 epigenetic silencing may reverse the inhibitory effects of estrogens and synergize with anti-estrogens, contributing to the overall better prognosis of Er-positive breast tumors. On the other side in TNBC, KEAP1 epigenetic silencing may results in a more aggressive phenotype by promoting cell survival and inhibiting apoptosis.
Chemotherapy plays an important role in the treatment of breast cancer. In a recent meta analysis reported by the Early Breast Cancer Trialists’ Collaborative Group, adjuvant treatment with polichemotherapy regimens determined a 25% proportional reduction in breast cancer mortality rate with an absolute reductions in breast cancer mortality of 6.5% at 10 y.Citation34 Despite this significant progress in the treatment of breast cancer, resistance to chemotherapeutic agents remains a critical issue in the therapeutical management of breast cancer patients. In vitro studies have demonstrated that KEAP1 somatic mutations are able to confer resistance to several anticancer drugs.Citation8,Citation11,Citation13,Citation15,Citation23 However, conflicting data are reported in the few studies analyzing the association between dysfunctional Nrf2/Keap1 interaction, patients survival and treatments.Citation15,Citation19,Citation25,Citation26 The standard treatment for both newly diagnosed and recurrent breast cancer is based on anthracyclines (epirubicin and vinorelbine) in polichemotherapy with cyclophosphamide, 5-fluorouracyl and, more recently, taxanes. In our study, KEAP1 promoter methylation was associated in both univariable and multivariable analyses with a better overall survival in the patients group receiving sequential therapy with antracyclines and cyclophosphamide followed by taxanes (EC/D), whereas no association was found in the group receiving antracyclines and cyclophosphamide. These results suggest that reduced Keap1 expression might be associated with a better response to chemotherapeutical regimens containing taxanes. However, further studies on larger patients cohorts homogenously treated will be needed to clarify the predictive role of KEAP1 genetic and epigenetic changes in breast cancer as well as in other tumor types.
In summary, we show that the Nrf2-negative regulator KEAP1 is frequently methylated in breast cancer. Moreover methylation is an early event in breast carcinogenesis suggesting that Keap1 reduced expression may play a role in the development and progression of the disease. In our patients’ cohort, KEAP1 methylation was associated with worse prognosis in patients characterized by a triple-negative phenotype but was predictive of reduced risk of tumor relapse in patients treated with EC/D chemotherapy regimen.
Materials and Methods
Patients and samples
We analyzed 102 patients treated by surgery at the Breast Unit IRCCS Casa Sollievo della Sofferenza, San Giovanni Rotondo (FG). Upon receipt from surgery, tissue sample from the bulk of the tumor, and normal breast tissue at least 2 cm distant from cancer are immediately frozen in liquid nitrogen and stored at –80°C until used. Prior written and informed consent is obtained from each patient in accordance with Institutional guidelines. For legal reasons only tumors greater than 1.0 cm can be sampled. As control, DNA sample extracted from 6 histologically confirmed normal breast tissues obtained from reductive mammoplasty were also analyzed.
Clinicopathological data are retrieved from patients’ charts and pathology reports. Pathological assessment includes evaluation of histological type, grade and stage. Estrogen receptor (Er), progesterone receptor (PgR), Ki67 labeling index and Her2 expression are evaluated by immunohistochemistry. Er and PgR immunoreactivity are reported according to ASCO/CAP guidelines and tumors with ≥ 1% of immunoreactivity are considered positive.Citation35 Her2 immunoreactivity assessment is performed according to the intensity and completeness of cell membrane staining. Only 3+ tumors are considered truly overexpressing Her2. Fluorescence in situ hybridization (FISH; PATHvision HER2 DNA probe kit, Abbott) is performed for tumors with a 2+ Her2 score by IHC and only cases with a FISH ratio (HER2 gene signals to chromosome 17 signals) ≥ 2 were considered amplified.Citation36
Bisulphite conversion
DNA was extracted from pre-invasive and invasive lesions for samples containing more than 70% of tumor cellsCitation37 and from normal breast tissues to be subjected to bisulphite treatment and purification using the Epitect Bisulfite kit (Qiagen Sci), according to manufacturer instructions. Bisulphite-modified DNA from the same treatment was used as template for fluorescence-based real-time Quantitative Methylation Specific PCR (QMSP).
Quantitative Methylation Specific PCR (QMSP)
KEAP1 methylation status was determined by using a QMSP assay that we previously validated by comparing methylation levels and bisulfite sequencing analysis of the same sample.Citation26 The KEAP1 primers/probe set was: forward 5′-TGCGGTCGTCGGATTACGAGGTCG-3′, reverse 5′CTTCCATCTCCCGATTTCGTTAC-3′; probe 5′-FAM-GTGGCGCGTAGTTTCGCGAG-TAMRA-3′ yielding a 111 bp amplicon. As reference gene a primers/probe set specific for a region of the ACTB gene not containing CpGs was used: forward 5′-TGGTGATGGAGGAGGTTTAGTAAGT-3′; reverse 5′-AACCAATAAAACCTACTCCTCCCTTAA-3′; probe 5′- FAM ACCACCACCCAACACACAATAACAAAC ACA TAMRA -3′, yielding an amplicon size of 133 bp.Citation26 Calibration curves for both target and reference genes were constructed using serial dilutions (90–0.009 ng) of a commercially available fully methylated DNA (CpGenome Universal Methylated DNA, Serologicals Corp.).
Amplification reactions were performed in triplicate in a volume of 10 μL that contained 50 ng bisulphite-modified DNA, 600 nM forward and reverse primers, 200 nM probe, 0.6 U of Platinum Taq polymerase (Life Technologies Corporation), 200 μM each of dATP, dCTP, dGTP, dTTP and 2 μl of PCR buffer.Citation26 PCR conditions were as follows: one step at 95°C for 3 min, 50 cycles at 95°C for 15 sec, and 60°C to 62°C for 1 min. PCR reactions were performed in 384-well plates on ABI PRISM 7900 Sequence detection system (Life Technologies Corporation) and were analyzed by SDS 2.4 software (Life Technologies Corporation). Each plate included calibration curves for the ACTB and KEAP1 genes, patients’ DNA samples, positive control (CpGenomeTM Universal Methylated DNA, Serologicals Corp.), and multiple water blanks. The relative level of methylated DNA was determined as a ratio of KEAP1 to ACTB and then multiplied by 1,000 for easier tabulation (average value of triplicates of gene of interest/average value of triplicates of ACTB × 1,000).
Mutation analysis
DNA obtained from the 20 invasive breast tumors was analyzed by fluorescence based direct sequencing for mutations in exons 4–7.Citation18 Amplification reactions were performed in a final reaction volume of 25 μL containing 100 ng of genomic DNA template, 0.25 nM dNTPs, 20 pmol of each primers, 1U HotMaster Taq polymerase (Eppendorf AG), in 1X PCR Reaction Buffer. PCR cycling conditions consisted of an initial denaturation step at 94°C for 2 min, followed by 35 cycles of 94°C for 30 sec, annealing for 30 sec, extension at 72°C for 30 sec, and ending with a final elongation step at 72°C for 7 min. PCR products were purified using GFX PCR DNA and Gel Band Purification Kit (GE Healthcare) and sequenced. Sequencing reactions were performed in 10 μL of final volume using 3 pmol of primer, 4–6 ng of DNA template and 1 μL of Big Dye Terminator Ready Reaction mix v. 1.1 (Life Technologies Corporation). Sequencing reactions were loaded on an ABI 3100 sequence detection system (Life Technologies Corporation) and analyzed using the Sequencing Analysis software v.3.7 (Life Technologies Corporation).
Statistical methods
Patients’ baseline characteristics were reported as median and interquartile range (IQR) or frequencies and percentages for continuous and categorical variables, respectively. Baseline comparisons were made using the Mann-Whitney U-test for continuous variables and Chi-square test for categorical variables. The overall differences between paired samples was assessed by the Skillings and Mack test Citation38, a generalization of Friedman test for unbalanced design which also allowed for post-hoc pairwise comparisons generalizing the Wilcoxon signed rank test. The discriminatory power of KEAP1 was assessed by estimating the area under the receiver operating characteristic (ROC) curve, using methylation levels in normal breast tissues and tumor samples. The optimal cut-off was assessed maximizing jointly sensitivity and specificity. Sensitivity and specificity, computed at the optimal cut-off, were reported along with their 95% confidence intervals (95% CI). The area under the ROC curve (AUC) was also reported along with its 95% CI. Samples were classified as methylated (M) or unmethylated (UM) based on whether KEAP1 to ACTB ratios were respectively above or below cut of values.
Time-to-event analysis was performed by univariable and multivariable COX proportional hazards regression models. Risks were reported as hazards ratios (HR) along with their 95% CI.
Time to progression was defined as the time between surgery and the first progression event. Overall Survival was defined as the time between surgery and death.
Furthermore, progression-free survival curves were estimated using Kaplan-Meier method and were graphically reported.
Furthermore, to evaluate interactions between KEAP1 and Er, PgR and HER2 status, and to identify distinct and homogeneous subgroups of patients in terms of overall survival, the RECPAM method was used.Citation39,Citation40 This tree-based method integrates the advantages of main effects COX regression and tree-growing techniques. At each partitioning step the method chooses the covariate and its best binary split to maximize the difference in the outcome of interest. The algorithm stops when user defined conditions (stopping rules) are met. To obtain more robust and stable split, a permutation approach was adopted to choose the best splitting variable.
A p value < 0.05 was considered for statistical significance. All analyses were performed using SAS Release 9.1.3 (SAS Institute). For the RECPAM analysis we used an SAS macro routine written by one of the authors (F.P.).
Additional material
Download Zip (184.2 KB)Acknowledgments
This study was supported by the “Ricerca Corrente 2012” funding granted by the Italian Ministry of Health and by the “5 × 1,000” voluntary contributions” and partially funded by the “Progetto Operativo Nazionale, PON 2011-2014 VIRTUALAB (PON01_01297), “Reti Laboratori pubblici di Ricerca: Progetto BioBOP” and AIRC Investigator Grant 2005–2007.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Kensler TW, Wakabayashi N, Biswal S. Cell survival responses to environmental stresses via the Keap1-Nrf2-ARE pathway. Annu Rev Pharmacol Toxicol 2007; 47:89 - 116; http://dx.doi.org/10.1146/annurev.pharmtox.46.120604.141046; PMID: 16968214
- Zhang DD. Mechanistic studies of the Nrf2-Keap1 signaling pathway. Drug Metab Rev 2006; 38:769 - 89; http://dx.doi.org/10.1080/03602530600971974; PMID: 17145701
- Shen G, Kong AN. Nrf2 plays an important role in coordinated regulation of Phase II drug metabolism enzymes and Phase III drug transporters. Biopharm Drug Dispos 2009; 30:345 - 55; http://dx.doi.org/10.1002/bdd.680; PMID: 19725016
- MacLeod AK, McMahon M, Plummer SM, Higgins LG, Penning TM, Igarashi K, et al. Characterization of the cancer chemopreventive NRF2-dependent gene battery in human keratinocytes: demonstration that the KEAP1-NRF2 pathway, and not the BACH1-NRF2 pathway, controls cytoprotection against electrophiles as well as redox-cycling compounds. Carcinogenesis 2009; 30:1571 - 80; http://dx.doi.org/10.1093/carcin/bgp176; PMID: 19608619
- Hayes JD, McMahon M. NRF2 and KEAP1 mutations: permanent activation of an adaptive response in cancer. Trends Biochem Sci 2009; 34:176 - 88; http://dx.doi.org/10.1016/j.tibs.2008.12.008; PMID: 19321346
- Padmanabhan B, Tong KI, Ohta T, Nakamura Y, Scharlock M, Ohtsuji M, et al. Structural basis for defects of Keap1 activity provoked by its point mutations in lung cancer. Mol Cell 2006; 21:689 - 700; http://dx.doi.org/10.1016/j.molcel.2006.01.013; PMID: 16507366
- Singh A, Misra V, Thimmulappa RK, Lee H, Ames S, Hoque MO, et al. Dysfunctional KEAP1-NRF2 interaction in non-small-cell lung cancer. PLoS Med 2006; 3:e420; http://dx.doi.org/10.1371/journal.pmed.0030420; PMID: 17020408
- Ohta T, Iijima K, Miyamoto M, Nakahara I, Tanaka H, Ohtsuji M, et al. Loss of Keap1 function activates Nrf2 and provides advantages for lung cancer cell growth. Cancer Res 2008; 68:1303 - 9; http://dx.doi.org/10.1158/0008-5472.CAN-07-5003; PMID: 18316592
- Lee DF, Kuo HP, Liu M, Chou CK, Xia W, Du Y, et al. KEAP1 E3 ligase-mediated downregulation of NF-kappaB signaling by targeting IKKbeta. Mol Cell 2009; 36:131 - 40; http://dx.doi.org/10.1016/j.molcel.2009.07.025; PMID: 19818716
- Tian H, Zhang B, Di J, Jiang G, Chen F, Li H, et al. Keap1: one stone kills three birds Nrf2, IKKβ and Bcl-2/Bcl-xL. Cancer Lett 2012; 325:26 - 34; http://dx.doi.org/10.1016/j.canlet.2012.06.007; PMID: 22743616
- Niture SK, Jaiswal AK. INrf2 (Keap1) targets Bcl-2 degradation and controls cellular apoptosis. Cell Death Differ 2011; 18:439 - 51; http://dx.doi.org/10.1038/cdd.2010.114; PMID: 20865015
- Niture SK, Jaiswal AK. Inhibitor of Nrf2 (INrf2 or Keap1) protein degrades Bcl-xL via phosphoglycerate mutase 5 and controls cellular apoptosis. J Biol Chem 2011; 286:44542 - 56; http://dx.doi.org/10.1074/jbc.M111.275073; PMID: 22072718
- Shibata T, Kokubu A, Gotoh M, Ojima H, Ohta T, Yamamoto M, et al. Genetic alteration of Keap1 confers constitutive Nrf2 activation and resistance to chemotherapy in gallbladder cancer. Gastroenterology 2008; 135:1358 - 68, 1368, e1-4; http://dx.doi.org/10.1053/j.gastro.2008.06.082; PMID: 18692501
- Yoo NJKH, Kim HR, Kim YR, An CH, Lee SH. Somatic mutations of the KEAP1 gene in common solid cancers. Histopathology 2012; 60:943 - 52; http://dx.doi.org/10.1111/j.1365-2559.2012.04178.x; PMID: 22348534
- Konstantinopoulos PA, Spentzos D, Fountzilas E, Francoeur N, Sanisetty S, Grammatikos AP, et al. Keap1 mutations and Nrf2 pathway activation in epithelial ovarian cancer. Cancer Res 2011; 71:5081 - 9; http://dx.doi.org/10.1158/0008-5472.CAN-10-4668; PMID: 21676886
- Hammerman PS, Hayes DN, Wilkerson MD, Schultz N, Bose R, Chu A, et al, Cancer Genome Atlas Research Network. Comprehensive genomic characterization of squamous cell lung cancers. Nature 2012; 489:519 - 25; http://dx.doi.org/10.1038/nature11404; PMID: 22960745
- Yoo NJ, Kim HR, Kim YR, An CH, Lee SH. Somatic mutations of the KEAP1 gene in common solid cancers. Histopathology 2012; 60:943 - 52; http://dx.doi.org/10.1111/j.1365-2559.2012.04178.x; PMID: 22348534
- Muscarella LA, Parrella P, D’Alessandro V, la Torre A, Barbano R, Fontana A, et al. Frequent epigenetics inactivation of KEAP1 gene in non-small cell lung cancer. Epigenetics 2011; 6:710 - 9; http://dx.doi.org/10.4161/epi.6.6.15773; PMID: 21610322
- Solis LM, Behrens C, Dong W, Suraokar M, Ozburn NC, Moran CA, et al. Nrf2 and Keap1 abnormalities in non-small cell lung carcinoma and association with clinicopathologic features. Clin Cancer Res 2010; 16:3743 - 53; http://dx.doi.org/10.1158/1078-0432.CCR-09-3352; PMID: 20534738
- Takahashi T, Sonobe M, Menju T, Nakayama E, Mino N, Iwakiri S, et al. Mutations in Keap1 are a potential prognostic factor in resected non-small cell lung cancer. J Surg Oncol 2010; 101:500 - 6; PMID: 20213688
- Wong TF, Yoshinaga K, Monma Y, Ito K, Niikura H, Nagase S, et al. Association of keap1 and nrf2 genetic mutations and polymorphisms with endometrioid endometrial adenocarcinoma survival. Int J Gynecol Cancer 2011; 21:1428 - 35; http://dx.doi.org/10.1097/IGC.0b013e31822d0eb2; PMID: 21897267
- Nioi P, Nguyen T. A mutation of Keap1 found in breast cancer impairs its ability to repress Nrf2 activity. Biochem Biophys Res Commun 2007; 362:816 - 21; http://dx.doi.org/10.1016/j.bbrc.2007.08.051; PMID: 17822677
- Jiang T, Chen N, Zhao F, Wang XJ, Kong B, Zheng W, et al. High levels of Nrf2 determine chemoresistance in type II endometrial cancer. Cancer Res 2010; 70:5486 - 96; http://dx.doi.org/10.1158/0008-5472.CAN-10-0713; PMID: 20530669
- Karihtala P, Kauppila S, Soini Y, Arja-Jukkola-Vuorinen. Oxidative stress and counteracting mechanisms in hormone receptor positive, triple-negative and basal-like breast carcinomas. BMC Cancer 2011; 11:262; http://dx.doi.org/10.1186/1471-2407-11-262; PMID: 21693047
- Ronkainen H, Vaarala MH, Kauppila S, Soini Y, Paavonen TK, Rask J, et al. Increased BTB-Kelch type substrate adaptor protein immunoreactivity associates with advanced stage and poor differentiation in renal cell carcinoma. Oncol Rep 2009; 21:1519 - 23; PMID: 19424632
- Muscarella LA, Barbano R, D’Angelo V, Copetti M, Coco M, Balsamo T, et al. Regulation of KEAP1 expression by promoter methylation in malignant gliomas and association with patient’s outcome. Epigenetics 2011; 6:317 - 25; http://dx.doi.org/10.4161/epi.6.3.14408; PMID: 21173573
- Hanada N, Takahata T, Zhou Q, Ye X, Sun R, Itoh J, et al. Methylation of the KEAP1 gene promoter region in human colorectal cancer. BMC Cancer 2012; 12:66; http://dx.doi.org/10.1186/1471-2407-12-66; PMID: 22325485
- Wang R, An J, Ji F, Jiao H, Sun H, Zhou D. Hypermethylation of the Keap1 gene in human lung cancer cell lines and lung cancer tissues. Biochem Biophys Res Commun 2008; 373:151 - 4; http://dx.doi.org/10.1016/j.bbrc.2008.06.004; PMID: 18555005
- Yager JD, Davidson NE. Estrogen carcinogenesis in breast cancer. N Engl J Med 2006; 354:270 - 82; http://dx.doi.org/10.1056/NEJMra050776; PMID: 16421368
- Cavalieri EL, Stack DE, Devanesan PD, Todorovic R, Dwivedy I, Higginbotham S, et al. Molecular origin of cancer: catechol estrogen-3,4-quinones as endogenous tumor initiators. Proc Natl Acad Sci U S A 1997; 94:10937 - 42; http://dx.doi.org/10.1073/pnas.94.20.10937; PMID: 9380738
- Montano MM, Chaplin LJ, Deng H, Mesia-Vela S, Gaikwad N, Zahid M, et al. Protective roles of quinone reductase and tamoxifen against estrogen-induced mammary tumorigenesis. Oncogene 2007; 26:3587 - 90; http://dx.doi.org/10.1038/sj.onc.1210144; PMID: 17160017
- Sripathy SP, Chaplin LJ, Gaikwad NW, Rogan EG, Montano MM. hPMC2 is required for recruiting an ERbeta coactivator complex to mediate transcriptional upregulation of NQO1 and protection against oxidative DNA damage by tamoxifen. Oncogene 2008; 27:6376 - 84; http://dx.doi.org/10.1038/onc.2008.235; PMID: 18663360
- Yao Y, Brodie AM, Davidson NE, Kensler TW, Zhou Q. Inhibition of estrogen signaling activates the NRF2 pathway in breast cancer. Breast Cancer Res Treat 2010; 124:585 - 91; http://dx.doi.org/10.1007/s10549-010-1023-8; PMID: 20623181
- Peto R, Davies C, Godwin J, Gray R, Pan HC, Clarke M, et al, Early Breast Cancer Trialists’ Collaborative Group (EBCTCG). Comparisons between different polychemotherapy regimens for early breast cancer: meta-analyses of long-term outcome among 100,000 women in 123 randomised trials. Lancet 2012; 379:432 - 44; PMID: 22152853
- Hammond ME, Hayes DF, Dowsett M, Allred DC, Hagerty KL, Badve S, et al. American Society of Clinical Oncology/College Of American Pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer. J Clin Oncol 2010; 28:2784 - 95; http://dx.doi.org/10.1200/JCO.2009.25.6529; PMID: 20404251
- Wolff AC, Hammond ME, Schwartz JN, Hagerty KL, Allred DC, Cote RJ, et al, American Society of Clinical Oncology, College of American Pathologists. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for human epidermal growth factor receptor 2 testing in breast cancer. J Clin Oncol 2007; 25:118 - 45; http://dx.doi.org/10.1200/JCO.2006.09.2775; PMID: 17159189
- Parrella P, Poeta ML, Gallo AP, Prencipe M, Scintu M, Apicella A, et al. Nonrandom distribution of aberrant promoter methylation of cancer-related genes in sporadic breast tumors. Clin Cancer Res 2004; 10:5349 - 54; http://dx.doi.org/10.1158/1078-0432.CCR-04-0555; PMID: 15328171
- Skillings JH, Mack GA. On the use of a Friedman-type statistic in balanced and unbalanced block designs. Technometrics 1981; 23:171
- Ciampi A, Negassa A, Lou Z. Tree-structured prediction for censored survival data and the Cox model. J Clin Epidemiol 1995; 48:675 - 89; http://dx.doi.org/10.1016/0895-4356(94)00164-L; PMID: 7730923
- De Berardis G, Pellegrini F, Franciosi M, Belfiglio M, Di Nardo B, Greenfield S, et al, QuED Study Group. Clinical and psychological predictors of incidence of self-reported erectile dysfunction in patients with type 2 diabetes. J Urol 2007; 177:252 - 7; http://dx.doi.org/10.1016/j.juro.2006.08.102; PMID: 17162057