Abstract
DNA methylation gradiently changes with age and is likely to be involved in aging-related processes with subsequent phenotype changes and increased susceptibility to certain diseases. The Hutchinson-Gilford Progeria (HGP) and Werner Syndrome (WS) are two premature aging diseases showing features of common natural aging early in life. Mutations in the LMNA and WRN genes were associated to disease onset; however, for a subset of patients the underlying causative mechanisms remain elusive. We aimed to evaluate the role of epigenetic alteration on premature aging diseases by performing comprehensive DNA methylation profiling of HGP and WS patients. We observed profound changes in the DNA methylation landscapes of WRN and LMNA mutant patients, which were narrowed down to a set of aging related genes and processes. Although of low overall variance, non-mutant patients revealed differential DNA methylation at distinct loci. Hence, we propose DNA methylation to have an impact on premature aging diseases.
Introduction
Hutchinson-Gilford Progeria Syndrome (HGP; OMIM:176670) and Werner Syndrome (WS; OMIM:277700) are two distinct premature aging diseases defined by aging related phenotypes occurring early in life.Citation1 As naturally aged individuals, HGP and WS patients present hair graying, skin thinning, atherosclerosis and osteopenia, but also show unique features such as subcutaneous tissue loss or laryngeal atrophy (high voice). Genetically, the disease could be traced back to mutations in lamin A (LMNA)Citation2,Citation3 and the Werner syndrome RecQ helicase like (WRN)Citation4 genes. However, the downstream causal events still remain elusive. In addition, a number of patients presenting a HGP or WS phenotype do not present any known genetic aberrations.
Herein, epigenetics could provide additional information by adding a level of gene regulation independent from genetic alterations. Epigenetics, understood in general terms as the inheritance of genome activity that does not depend on the strict DNA sequence, is known to be dynamic and to adapt to the surrounding circumstances.Citation5 In addition, epigenetics provides an explanation for the phenotypic differences of genetically identical beings, such as the examples of monozygotic twins,Citation6,Citation7 cloned animalsCitation8 or the Agouti mice.Citation9,Citation10 To address the issue of whether epigenetic variants can be associated with the disease phenotypes of premature aged patients, we analyzed the DNA methylation (the most recognized and studied epigenetic mark) profiles of HGP and WS samples. We approach both diseases from an epigenetic point of view assuming missregulation to take place beyond the genetic blueprint.
Results and Discussion
We performed the Infinium DNA methylation BeadChip platform (Illumina), analyzing more than 450,000 CpG sites genome-wide. DNA was extracted from Epstein-Barr virus (EBV) immortalized B-cells (lymphoblastoid cell lines; LCLs) obtained from two WRN (AG07896, AG11385) and one LMNA (AG19911) mutant WS patient ().Citation11 In addition, we analyzed one non-mutant WS patient (AG03364) and three non-mutant HGP patients. It is of note that the non-mutant HGP samples consist of direct relatives, represented by a father (AG15693) and its two daughters (AG15694, AG15695). The DNA methylation data are freely available at the Gene Expression Omnibus (GEO) database: www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=xlcjfieweoacwna&acc=GSE42865.
Table 1. Patient and healthy samples analyzed on the Infinium DNA methylation BeadChip
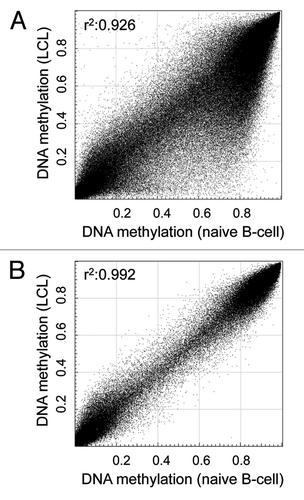
Noticing an effect of the EBV immortalization on the epigenome of the analyzed control LCL samples, as illustrated by absolute DNA methylation levels of naive CD19+ B-cells and a LCL, ), we initially filtered for CpG sites without variability between healthy LCLs (n = 3) and healthy naive B-cells (n = 3) (δ average β-value < 0.05). To further exclude an impact of blood cell composition of the analyzed LCL samples, especially important when comparing LCLs and peripheral blood mononuclear cells (PBMC) data from a previous aging study,Citation12 we additionally excluded CpG sites revealing variability between naive B-cells (n = 3) and samples obtained from PBMC (n = 3; δ average β-value < 0.05). After further exclusion of CpG sites on gender chromosomes and those containing single nucleotide polymorphisms in their detection probe or interrogated CpG site (Caucasian population, 1000 Genome Project, > 1% frequency) and poor detection quality (detection p value > 0.01), we analyzed 272,290 CpG sites in the here presented study. Following filtering for probes detecting here unmodelled sources, we observed a very high correlation (R2 > 0.99) among all analyzed healthy control samples, as exemplary illustrated in .
Figure 1. Probe filtering results in consistent DNA methylation profiles between naive and immortalized samples. (A) DNA methylation level of 448,376 CpG sites (after filtering for high quality autosomal probes not overlapping SNPs) of a representative lymphoblastoid cell line (LCL) and a naive B-cell sample. (B) DNA methylation levels of 272,290 CpG sites of the same samples after excluding probes affected by Epstein-Barr virus immortalization and cell composition differences. The correlation coefficient (r2) was calculated using Pearson’s correlation analysis.
Mutant patients exhibit global DNA methylation differences
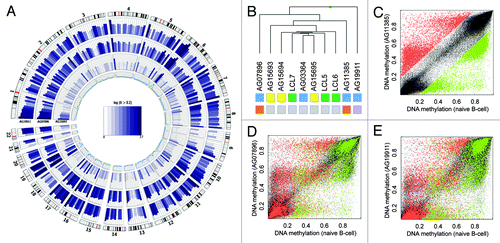
Genome-wide DNA methylation variation () and unsupervised clustering analysis () of the DNA methylation profiles of HGP and WS patient DNA revealed profound differences between mutant and non-mutant samples. The WRN and LMNA mutant samples clearly clustered separately from the controls, suggesting mutation-specific DNA methylation profiles with possible causality on disease onset. Single samples comparisons revealed globally similar DNA methylation profiles with a fraction of CpG sites gaining methylation (in a healthy unmethylated context) and loosing methylation in previous hypermethylated regions (). Importantly, we noticed an overlap of differentially methylated CpG sites. However, each sample revealed additional unique alterations ().
Figure 2. Mutant patients exhibit global DNA methylation differences. (A) Genome-wide DNA methylation changes of a non-mutant (AG15693), a WRN gene mutant (AG07896, AG11385) and a LMNA gene mutant (AG19911) premature aging patient compared with a healthy donor (LCL6) and displayed by Circos representation.Citation21 Displayed in color-code are the number of differentially methylated CpG site (δ average β-value > 0.2) in windows of 10 Mbp width. The inner circle represents the total number of analyzed CpG sites in the respective window (0–7,000). (B) Unsupervised hierarchical clustering of healthy donors (green) or HGP (yellow) and WS (blue) patient samples. Gene mutations are indicated (WRN: orange, LMNA, purple; non-mutant: gray). (C) Absolute DNA methylation levels of AG11385 (WRN mutant) and LCL6 (healthy donor). Highlighted are hypomethylated (δ < -0.2, green) and hypermethylated (δ > 0.2, red) CpG sites in respect the healthy control (LCL6). (E) Absolute DNA methylation level of AG07896 (WRN mutant) and LCL6 (healthy donor). Highlighted are hypomethylated (green) and hypermethylated (red) CpG sites of AG11385. (D) Absolute DNA methylation level of AG19911 (LMNA mutant) and LCL6 (healthy donor). Highlighted are hypomethylated (green) and hypermethylated (red) CpG sites of AG11385.
The non-mutant WS and HGP samples presented DNA methylation profiles that were poorly distinguishable between healthy and diseased samples (). Interestingly, the non-mutant sample of the father (AG15693) revealed an increase of variability compared with the two daughters (AG15694, AG15695), likely reflecting the fact that, besides presenting the HGP phenotype, the father was 28 y older at sample acquisition.
Differentially methylated sites in mutant patients
In order to extract particular disease-related differences between controls and samples obtained from premature aging patients, we determined differentially methylated CpG sites (dmCpGs) between the controls (n = 3) and diseased samples (δ average β-value > 0.2).
Initially, we were interested in epigenetic alteration associated to the disease-related gene mutations of LMNA or WRN. As mutations in LMNA are causing both HGP and WS, whereas WRN mutations are exclusively related to the latter, we analyzed both mutations separately. We observed that the mutant samples presented a number of overlapping alterations; however, there was a high proportion of sample-specific alteration () and inter-sample variation (). Using the aforementioned selection criteria, we determined 3,544 consistent differentially methylated CpG sites associated to mutation in the WRN gene (13,501 for AG11385 and 17,506 for AG07896). Although affected by the same genetic defect the WRN mutant samples revealed a high variability, which might be explained by the nature of the genetic defect. The Werner syndrome RecQ helicase gene is involved in DNA replication, recombination and DNA repair (3′ to 5′ exonuclease activity). Impaired function of WRN eventually results in double strand breaks and accumulation of mutations. The here observed epimutations could be a direct consequence of mutant CpG sites or CpG methylation-associated sequences in their proximity (methylation quantitative trait lociCitation13). Considering the stochastic distribution of such events, which is supported by the equal genome-wide distribution (), we expected a few driver events masked by a mass of passenger events on genetic and epigenetic level. To address this critical point, we assumed that, due to the phenotype similarities to normal aging, the disease causing driver events were also detectable in natural aged samples. Consequently, we integrated DNA methylation data obtained from a set of newborn and centenarian samples previously analyzed in our laboratory for aging specific changes.Citation12 Here, 144 CpG showed differential methylation in the natural aged and in the WRN mutant sample sets and thus represent potential driver candidates for the premature aging phenotype of the WS patients (Table S1). These sites include gene promoters involved and significantly enriched in I-kappaB kinase/NF-kappaB signaling (CASP8, IL1RL1, LGALS1) and the proteinaceous extracellular matrix formation (ADAMTS4, LGALS1, PODNL1, ZP3) (Fisher’s exact test: p < 0.01). All of these candidates are likely to be involved in phenotypic changes observed during aging.
While patients with WRN mutation have to cope with an accumulation of DNA damage, LMNA mutant cells are associated to severe structural alterations in the nucleus, leading to chromatin reorganization and subsequent epigenetic alterations. Therefore, most of the detected variation in DNA methylation will be passenger events, not directly related to disease onset. Consequently, we applied the aforementioned selection criteria and comparisons to define dmCpGs for the WS patient harboring a LMNA mutation. Here, we identified 18,480 dmCpGs, of which 485 also revealed differential methylation in the natural aging data set (Table S2). Interestingly, the NF-kappaB nucleus import mechanism was a significantly enriched ontology term among the differentially methylated genes (Fisher’s exact test: p < 0.01; PRKCG, NLRP12), suggesting this inflammation-related process plays a crucial role in the aging process of healthy and diseased individuals.
Non-mutant patients revealed consistent changes in DNA methylation
Analyzing non-mutant premature aging samples, we took advantage of the unique properties of the three HGP family members. Assuming a similar genetic background of the samples, we aimed to extract common dmCpGs and suggested those to present a particular association to the premature aging disease. Here, we observed 78 sites with consistent changes in DNA methylation (Table S3). Most strikingly, the promoter of the non-coding RNA LOC149837 harbored four dmCpGs in the close proximity of the transcription start site (-51 bp), forming a differential methylated region with high potential as an important factor for gene regulation (). LOC149837, also annotated as LINC00654, is defined as long intergenic non-protein coding RNA (lincRNA) with to date unknown biological function. Interestingly, the transcript is located in a cluster of PIWI-associated RNAs (piRNAs) (piRNA bank ID: 103; chr20:5461000–5500000), suggesting the region is a so far poorly recognized piRNA host transcript with association to premature aging. The approximately 30 nt-sized small RNAs participate in germline-specific regulatory processes, including silencing of transposable elements by direct interaction or epigenetic repression.Citation14 As a subset of piRNAs, including the here identified, do not overlap transposable sequences, an additional function of the small RNAs is suspected. As the epimutation of LOC149837 is present in all seven analyzed premature aged samples (including the mutation driven phenotypes, ), we suggest the differential promoter hypomethylation to be a common downstream event with crucial function in disease onset. We further propose the inappropriate promoter hypomethylation of LOC149837 and subsequent piRNA activation as causative epigenetic event for the development of HGP and WS.
Figure 3. Candidate genes for premature aging phenotypes. (A) Absolute DNA methylation levels of CpG sites in the promoter region of LOC149837 of non-mutant patients (AG) and healthy donors (LCL). The distance to the transcription start site (TSS) is indicated. (B) DNA methylation levels (low: green; high: red) of LOC149837 promoter CpG sites in all analyzed premature aging patients (AG) and healthy donors (LCL). The cluster was calculated using Manhattan distances. (C) Absolute DNA methylation level of CpG sites in the promoter region of POLD3 of non-mutant patients (AG) and healthy donors (LCL). The distance to the transcription start site (TSS) is indicated.
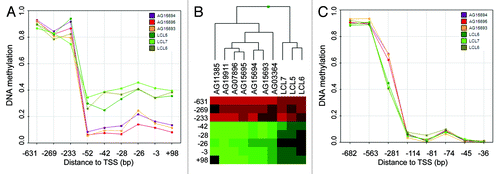
In order to determine yet unidentified disease-related genetic alteration in addition to the previously identified (WRN and LMNA), we screened differentially methylated gene promoters of the non-mutated samples for common regulatory mechanisms. Here, the 25 transcripts associated to disease-specific CpG sites were enriched for transcription factor binding sites for MYB (z-test; TRANSFAC: FDR < 0.05), a transcription factor associated to aging,Citation15 including the response to oxidative stress of aged cells.Citation16 It is of note that the DNA polymerase POLD3 is among the MYB targets, whose deregulation, due to its DNA repair activity, might have a direct impact on the disease phenotypes as it was observed for the DNA repair gene WRN (). The significant enrichment of MYB binding sites in differentially methylated gene promoters led us to hypothesize if the transcription factor itself is targeted by genetic alterations, eventually gene mutation, causing the HGP phenotype. Although speculative at this stage, these results might inspire further studies involving targeted sequencing of the MYB gene locus.
In summary, detecting differentially methylated CpG sites in potential disease-related genes and pathways, we hypothesize DNA methylation and epigenetic regulation to play an important role in disease onset of premature aging diseases, such as HGP and WS. With the transcription factor MYB and NF-kappaB associated pathways, we do not only suggest potentially novel susceptibility genes and signaling cascades, but we also present a yet poorly acknowledged way to approach the disease.
Materials and Methods
Sample description
The DNA of four Werner syndrome (WS) and three Hutchinson-Gilford Progeria (HGP) patients was obtained from Coriell Cell Repositories. The samples consisted of 2 WRN (AG07896, AG11385), 1 LMNA (AG19911) mutant and 4 patients without yet described alterations (AG03364), including 3 relatives with disease phenotype (AG15693, AG15694, AG15695). These samples had been collected and anonymized by National Institute of General Medical Science (NIGMS), and all subjects had provided written consent for their experimental use. Peripheral blood was obtained from healthy donors and peripheral blood mononuclear cells (PBMC) were extracted using a Ficoll gradient. To separate CD19 positive cells CD19 MicroBeads (Miltenyi Biotec) were applied following the manufacturer’s instructions. DNA was extracted using Phenol:Chloroform:Isoamylalcohol (Sigma). B-cells were immortalized by Epstein-Barr virus (EBV) applying previous published protocol.Citation17
Infinium HumanMethylation450 BeadChip
All DNA samples were assessed for integrity, quantity and purity by electrophoresis in a 1.3% agarose gel, picogreen quantification, and nanodrop measurements. All samples were randomly distributed into 96 well plates. Bisulfite conversion of 500 ng of genomic DNA was performed using EZ DNA methylation kit (Zymo Research) following manufacturer’s instructions. 200 ng of bisulfite converted DNA were used for hybridization on the HumanMethylation450 BeadChip (Illumina). Briefly, samples were whole genome amplified followed by an enzymatic end-point fragmentation, precipitation and resuspension. The resuspended samples were hybridized onto the beadchip for 16 h at 48°C and washed. A single nucleotide extension with labeled dideoxy-nucleotides was performed and repeated rounds of staining were applied with a combination of labeled antibodies differentiating between biotin and DNP.
Data were normalized using GenomeStudio V2010.3 (Illumina). The DNA methylation level is displayed as β-values ranging from 0 to 1. Methylation level (β-value) for each of the 485,577 CpG sites were calculated as the ratio of methylated signal divided by the sum of methylated and unmethylated signals plus 100. After normalization step, probes related to X and Y chromosomes were removed as well as those containing a SNPs with a frequency > 1% (Caucasian population; 1000 Genome project) in the probe sequence or interrogated CpG site. Samples were clustered by hierarchical clustering using Manhattan distances.
Transcription factor binding enrichment
Transcription factor enrichment analysis was performed using PSCAN.Citation18 Transcription factor binding annotations were extracted from TRANSFAC. The gene promoter region was defined as -1,000 and +0 bp to the transcription start site. Z-test p-values were corrected for multiple hypotheses testing using the Bonferroni method.
Functional enrichment analysis
Gene Ontology (GO) enrichment analysis was performed using the Database for Annotation, Visualization and Integrated Discovery (DAVID; v6.7).Citation19,Citation20
Additional material
Download Zip (153.4 KB)Acknowledgments
The research leading to these results has received funding from the European Research Council (ERC) grant EPINORC under the agreement n° 268626, the MICINN Project – SAF2011-22803, the Cellex Foundation, the European Community's Seventh Framework Programme (FP7/2007-2013) by the grant HEALTH-F5-2011-282510 - BLUEPRINT and the Health and Science Departments of the Generalitat de Catalunya. M.E. is an ICREA Research Professor.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Hegele RA. Drawing the line in progeria syndromes. Lancet 2003; 362:416 - 7; http://dx.doi.org/10.1016/S0140-6736(03)14097-4; PMID: 12927424
- Eriksson M, Brown WT, Gordon LB, Glynn MW, Singer J, Scott L, et al. Recurrent de novo point mutations in lamin A cause Hutchinson-Gilford progeria syndrome. Nature 2003; 423:293 - 8; http://dx.doi.org/10.1038/nature01629; PMID: 12714972
- De Sandre-Giovannoli A, Bernard R, Cau P, Navarro C, Amiel J, Boccaccio I, et al. Lamin a truncation in Hutchinson-Gilford progeria. Science 2003; 300:2055; http://dx.doi.org/10.1126/science.1084125; PMID: 12702809
- Yu CE, Oshima J, Fu YH, Wijsman EM, Hisama F, Alisch R, et al. Positional cloning of the Werner’s syndrome gene. Science 1996; 272:258 - 62; http://dx.doi.org/10.1126/science.272.5259.258; PMID: 8602509
- Feinberg AP. Phenotypic plasticity and the epigenetics of human disease. Nature 2007; 447:433 - 40; http://dx.doi.org/10.1038/nature05919; PMID: 17522677
- Kaminsky ZA, Tang T, Wang S-C, Ptak C, Oh GHT, Wong AHC, et al. DNA methylation profiles in monozygotic and dizygotic twins. Nat Genet 2009; 41:240 - 5; http://dx.doi.org/10.1038/ng.286; PMID: 19151718
- Fraga MF, Ballestar E, Paz MF, Ropero S, Setien F, Ballestar ML, et al. Epigenetic differences arise during the lifetime of monozygotic twins. Proc Natl Acad Sci U S A 2005; 102:10604 - 9; http://dx.doi.org/10.1073/pnas.0500398102; PMID: 16009939
- Rideout WM 3rd, Coetzee GA, Olumi AF, Jones PA. 5-Methylcytosine as an endogenous mutagen in the human LDL receptor and p53 genes. Science 1990; 249:1288 - 90; http://dx.doi.org/10.1126/science.1697983; PMID: 1697983
- Michaud EJ, van Vugt MJ, Bultman SJ, Sweet HO, Davisson MT, Woychik RP. Differential expression of a new dominant agouti allele (Aiapy) is correlated with methylation state and is influenced by parental lineage. Genes Dev 1994; 8:1463 - 72; http://dx.doi.org/10.1101/gad.8.12.1463; PMID: 7926745
- Waterland RA, Jirtle RL. Transposable elements: targets for early nutritional effects on epigenetic gene regulation. Mol Cell Biol 2003; 23:5293 - 300; http://dx.doi.org/10.1128/MCB.23.15.5293-5300.2003; PMID: 12861015
- Sandoval J, Heyn H, Moran S, Serra-Musach J, Pujana MA, Bibikova M, et al. Validation of a DNA methylation microarray for 450,000 CpG sites in the human genome. Epigenetics 2011; 6:692 - 702; http://dx.doi.org/10.4161/epi.6.6.16196; PMID: 21593595
- Heyn H, Li N, Ferreira HJ, Moran S, Pisano DG, Gomez A, et al. Distinct DNA methylomes of newborns and centenarians. Proc Natl Acad Sci U S A 2012; 109:10522 - 7; http://dx.doi.org/10.1073/pnas.1120658109; PMID: 22689993
- Bell JT, Pai AA, Pickrell JK, Gaffney DJ, Pique-Regi R, Degner JF, et al. DNA methylation patterns associate with genetic and gene expression variation in HapMap cell lines. Genome Biol 2011; 12:R10; http://dx.doi.org/10.1186/gb-2011-12-1-r10; PMID: 21251332
- Siomi MC, Sato K, Pezic D, Aravin AA. PIWI-interacting small RNAs: the vanguard of genome defence. Nat Rev Mol Cell Biol 2011; 12:246 - 58; http://dx.doi.org/10.1038/nrm3089; PMID: 21427766
- Hwang IK, Moon SM, Yoo K-Y, Li H, Kwon HD, Hwang HS, et al. c-Myb immunoreactivity, protein and mRNA levels significantly increase in the aged hippocampus proper in gerbils. Neurochem Res 2007; 32:1091 - 7; http://dx.doi.org/10.1007/s11064-006-9278-5; PMID: 17401667
- Lee Y-H, Lee N-H, Bhattarai G, Hwang P-H, Kim T-I, Jhee E-C, et al. c-myb has a character of oxidative stress resistance in aged human diploid fibroblasts: regulates SAPK/JNK and Hsp60 pathway consequently. Biogerontology 2010; 11:267 - 74; http://dx.doi.org/10.1007/s10522-009-9244-0; PMID: 19707884
- Miller G, Lipman M. Release of infectious Epstein-Barr virus by transformed marmoset leukocytes. Proc Natl Acad Sci U S A 1973; 70:190 - 4; http://dx.doi.org/10.1073/pnas.70.1.190; PMID: 4346033
- Zambelli F, Pesole G, Pavesi G. Pscan: finding over-represented transcription factor binding site motifs in sequences from co-regulated or co-expressed genes. Nucleic Acids Res 2009; 37:Web Server issue W247-52; http://dx.doi.org/10.1093/nar/gkp464; PMID: 19487240
- Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009; 4:44 - 57; http://dx.doi.org/10.1038/nprot.2008.211; PMID: 19131956
- Huang W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 2009; 37:1 - 13; http://dx.doi.org/10.1093/nar/gkn923; PMID: 19033363
- Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, et al. Circos: an information aesthetic for comparative genomics. Genome Res 2009; 19:1639 - 45; http://dx.doi.org/10.1101/gr.092759.109; PMID: 19541911