Abstract
The gene Oct4 encodes a transcription factor critical for the maintenance of pluripotency and self-renewal in embryonic stem cells. In addition, improper re-activation of Oct4 contributes to oncogenic processes. Herein, we describe a novel designer zinc finger protein (ZFP) capable of upregulating the endogenous Oct4 promoter in a panel of breast and ovarian cell lines carrying a silenced gene. In some ovarian tumor lines, the ZFP triggered a strong reactivation of Oct4, with levels of expression comparable with exogenous Oct4 cDNA delivery. Surprisingly, the reactivation of Oct4 required a KRAB domain for effective upregulation of the endogenous gene. While KRAB-containing ZFPs are traditionally described as transcriptional repressors, our results suggest that these proteins could, in certain genomic contexts, function as potent activators and, thus, outline an emerging novel function of KRAB-ZFPs. In addition, we document a novel ZFP that could be used for the epigenetic reprograming of cancer cells.
Introduction
In recent years, extensive research activity has been devoted to the study of pluripotency transcription factors (TFs). Oct4 (POU5F1) encodes a homeodomain-containing TF essential for the establishment and maintenance of self-renewal and pluripotency.Citation1 Oct4 binds specific sequences of DNA through cooperative interactions with other TFs, such as Sox2. These TF-TF interactions provide context-dependent combinatorial opportunities and binding epitopes for activator complexes, repressor complexes and chromatin remodelers.Citation2-Citation5 Genes upregulated by Oct4 include transcriptional partners involved in self-renewal, such as Sox2 and Nanog. In embryonic stem cells (ESCs), the TFs Oct4, Sox2 and Nanog form a feed forward auto-regulatory feedback loop, which results in activation of self-renewal, while repressing differentiation gene programs.Citation6,Citation7
Differentiation of ESCs is critically controlled by epigenetic mechanisms, notably DNA methylation and histone methylation (H3K9me). Incorporation of DNA methylation in the Oct4 promoter, catalyzed by de novo methyltransferases Dnmt3a and Dnmt3b, is considered a critical and “irreversible” epigenetic switch, which forces ESCs to differentiate.Citation8,Citation9 Hence, the vast majority of somatic cells, such as fibroblasts, retain a methylated and silenced Oct4 promoter. In contrast, this gene has been reported to be expressed in some stem cells, such as ESCs, stem cells of the germinal line,Citation10 and more recently, breast-milk-derived stem cells.Citation11 In addition, overexpression of Oct4 has been associated with oncogenic processes in several malignancies, such as breastCitation12 and ovarian cancers.Citation13,Citation14
Given the principal role of Oct4 silencing in differentiation, cell engineering and cancer, novel molecular strategies to revert this epigenetic state are of significant interest. The endogenous Oct4 promoter is commonly reactivated by exogenous delivery of specific cDNA combinations, including the Oct4, Sox2, Klf4 and c-Myc cDNAs, in a process referred as reprogramming to pluripotency.Citation15,Citation16 The crucial involvement of Oct4 in reprogramming to pluripotency was elegantly demonstrated by the generation of induced pluripotent cells by a single delivery of Oct4 cDNA in neural stem cells.Citation17 Moreover, it is well recognized that the bottleneck of the reprogramming process involves the re-activation and de-silencing of the endogenous pluripotency factors.Citation18 However, the exogenous expression of cDNAs often entails an incomplete erasure of the epigenetic memory of the cell of origin, resulting in incomplete reprogramming of CG methylation and other histone modifications, together with an aberrant epigenetic signature, which is transmitted in differentiated iPS cells.Citation19,Citation20 The erasure of epigenetic silencing marks can be facilitated by a combination of cDNAs and small molecules,Citation21 particularly chromatin remodeling agents, such as DNA methyltransferase, H3K9-methyltransferase and histone deacetylase (HDAC) inhibitors.Citation22,Citation23 In the last few years, several enzymatic activities associated with DNA demethylation have been identified in ESCs and these shown to play a role in early events of reprogramming.Citation24,Citation25 More recently, sequence-specific engineered proteins, Transcription Activator Like effectors (TALEs), have been designed to reactivate pluripotency genes, such as Sox222, Klf4Citation22 and Oct4,Citation26 in somatic cells. While Oct4-specific TALEs are not capable of upregulating the endogenous gene, combinations of TALEs with chromatin remodelers have been shown to reactivate the Oct4 promoter, outlining the importance of genomic silencing as an epigenetic blockade for artificial regulation.Citation26
In addition to TALEs, engineered Zinc Finger (ZF) proteins (ZFPs) or Artificial Transcription Factors (ATFs), represent an alternative strategy to modulate the epigenetic state of targeted promotersCitation27,Citation28. In contrast with TALEs, ZFPs are generated from mammalian SP1 backbones, which have the advantage to bind with high affinity CG rich sequences abundantly represented in proximal promoters.Citation29 Some designer ZFPs have been shown to reactivate the tumor suppressor gene mammary serine protease inhibitor (maspin) in cell lines carrying a high density of methylated CpG di-nucleotidesCitation30-Citation34. In addition to methylated tumor suppressors, 6ZF arrays targeting 18-base pair (bp) sites in promoters have been shown to bind and regulate a large spectrum of cellular targets. Moreover, like their natural counterparts, artificial ZFP require specific activators and repressor domains to promote differential expression of targeted genes.Citation35,Citation36 More recently, we have described nearly 95% repression of Sox2 via ZFP attached to a Kruppel associated Box (KRAB or SKD domainCitation37) and ~60% repression with the same ZFPs linked to a Dnmt3a catalytic domain,Citation38 in breast cancer cells. KRAB-containing ZFPs are traditionally regarded as potent transcriptional repressors via KAP-1-mediated recruitment of Histone deacetylase NuRD complex, Histone deacetylases (HDACs), histone methyltransferase SETDB1 and heterochromatin protein 1 (HP1).Citation39,Citation40 In addition to their role as transcriptional repressors, emerging work on natural ZFPs by genome-wide ChIP-Seq mapping suggest that at certain loci some KRAB-containing ZFPs, such as ZFN263,Citation41 could function to activate endogenous genes.
In this manuscript, we describe the construction of a designer ZFP composed of 6ZF domains, able to reactivate the endogenous Oct4 promoter in a panel of cell lines carrying a silenced gene. Intriguingly, only when the 6ZFs were linked to the KRAB domain was a potent upregulation of the endogenous gene achieved. The potency of the ZFP depended on the cell line analyzed, which suggests that the genomic microenvironment critically influences the regulatory outcome of KRAB-ZFPs. In some ovarian cancer cell lines, the activation of Oct4 mediated by the ZFP was similar to the levels of activation achieved by ectopic cDNA delivery. Our results indicate that ZFPs linked to KRAB domains can function, in specific chromatin contexts, as potent activators of silenced genes. Our studies support a new paradigm for the study of KRAB-ZFPs, and for the design of artificial activators in epigenetic reprogramming and cancer therapeutics.
Results
Oct4 silencing in a panel of breast and ovarian cancer samples
We first examined the levels of expression of Oct4 and two of its self-renewal TF targets, Sox2 and Nanog, in a panel of breast and ovarian cancer cell lines and tissue specimens by quantitative real-time PCR (qRT-PCR, and C). As expected, we found Oct4, Sox2 and Nanog silenced in primary cell lines such as human dermal fibroblasts, human mammary epithelial cells (HMECs) and non-transformed, immortalized breast epithelial cell lines, such the MCF-12A line. In contrast, all three TFs were abundantly expressed in hESCs and in the teratoma cell line NT2/D1, as expected from a positive auto-regulatory feedback loop reported between Oct4, Sox2 and Nanog in stem cellsCitation6 (). p86-OCBT-L1 is a breast cancer cell line generated by overexpression of hOct4 cDNA followed by serial passaging of breast epithelial cells from a mammoplastic donor (p86).Citation12 These cells acquired tumor-initiating features and re-activated the endogenous Oct4 and Nanog mRNAs,Citation12 and were thus used as a positive control in the cancer cell line panel.
Figure 1. Differential endogenous expression of Oct4, Sox2 and Nanog in breast and ovarian cell lines. Endogenous Oct4, Sox2 and Nanog mRNA levels as assessed by quantitative real-time PCR in a panel of different breast (A) and ovarian (B) cell lines. Real-time quantification of each gene expression was normalized to the fibroblast cell samples. hESCs, p86-OTBC-L1 and NT2/D1 cells represented positive controls for Oct4, Sox2 and Nanog expression. Primary ovarian cancer cell cultures were generated from cells isolated from the ascites of patients with epithelial ovarian cancer. Experiments were run in triplicate and data represent the mean ± SD of three independent experiments as determined by the Student’s t-test (*p ≤ 0.01). (C) Immunofluorescence (IF) Oct4 protein expression and intracellular localization of Oct4 in different human cell lines: Fibroblasts, Human Mammary Epithelial cells (HMEC) and cancer cells: MDA-MB-231 (Breast), OVCAR-3 and PA-1 (Ovarian) and p86-OTBC-L1 (breast) cell lines. (D) Methylation status of Oct4 promoter in Fibroblasts, HMEC, MDA-MB-231 OVCAR-3, PA-1 and p86-OCTB-L1 cell lines. The X-axis represent the nucleotide position relative to translation start site, and the Y-axis the percentage of methylation along the Oct4 proximal promoter, assessed with a Sequenom EpiTYPER platform. Pink arrows indicate specific CpG di-nucleotides where methylation frequencies decreased significantly in the tumor samples relative to the non-transformed fibroblast or HUMEC cells.
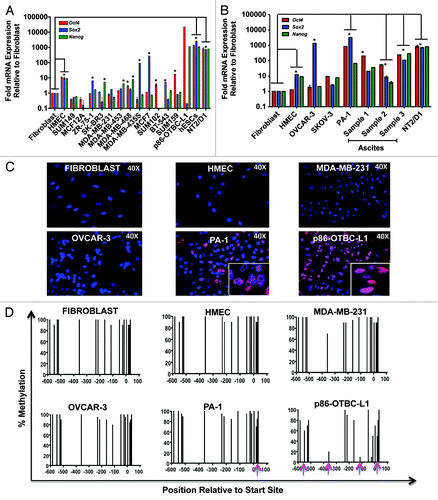
In breast and ovarian cancer samples, heterogeneous portraits of mRNA upregulation were observed, with no overall correlation between Oct4, Sox2 and Nanog transcript levels in the cancer cells. These data clearly demonstrated aberrant regulation of Oct4, Sox2 and Nanog mRNA expression in the tumor cells relative to somatic cell lines. Some breast cancer lines, such as MCF7, exhibited very high levels of Sox2, which were comparable with those observed in hESCs. In the vast majority of breast cancer lines examined both, Oct4 and Nanog mRNA levels were at least two orders of magnitude lower than hESCs and only ~10-fold higher than those detected in non-transformed cells. In contrast with breast cancer cell lines, some ovarian cancer lines and cell lines derived from ascites isolated from serous epithelial ovarian cancer (SOC) patients abundantly expressed Oct4, Sox2 and Nanog, with the highest expression of Oct4 detected in the PA-1 line (). The Oct4, Sox2 and Nanog mRNA expression was further validated in a panel of tissue specimens isolated from SOC patients, which revealed high levels of pluripotency TFs in some primary tumor tissue and in less extent in abdominal metastases isolated from the same patients (Fig. S1A). The expression of Oct4 in breast and ovarian cell lines and in some SOC tumor specimens was verified by immunofluorescence (IF) and immunohistochemistry (IHC), revealing predominantly a nuclear protein distribution. Moreover, some ovarian cancer cell lines, such as PA-1 and tumor specimens exhibited some weak cytoplasmic staining, which also has been reported by other groupsCitation14 (; Fig. S1B). The IF staining of Oct4 in the PA1 line revealed a high degree of heterogeneity, with high expression of the protein observed in a few cells in the population ().
In ESCs it has been demonstrated that epigenetic regulation of the Oct4 promoter, specifically DNA methylation, functions as an epigenetic switch, which forces stem cells to differentiate.Citation42 DNA methylation in the Oct4 promoter has been recognized as one of the main blockades for somatic cell reprogramming.Citation43,Citation44 Unlike somatic cells, such as fibroblasts, both ESCs and iPSCs carry a de-methylated Oct4 promoter, which is associated with high expression of the endogenous gene. We next studied DNA methylation frequencies in the Oct4 promoter, using a Sequenom EpiTYPER platform, in the panel of cancer cell lines to examine if silencing of Oct4 was also associated with differential DNA methylation in the tumor cells. In normal, non-transformed mammary epithelial cells, high levels of DNA methylation in the Oct4 promoter were observed, with similar frequencies to those reported in human dermal fibroblasts. Similarly to somatic cells, we found high levels of DNA methylation in tumor cell lines carrying silenced Oct4, such the MDA-MB-231 cell line. In addition, some CpG di-nucleotides were found differentially methylated in tumor cell lines having high levels of expression of Oct4, most notably, in the p86-OCBT-L1 line, which had an enriched Oct4-expressing cell population. However, the DNA methylation frequencies observed in the majority of tumor lines were much higher that those reported in ESCs and iPSCs.Citation15,Citation16,Citation18,Citation45 In summary, these data indicated that the vast majority of breast and ovarian cancers examined retained DNA methylation frequencies in the Oct4 promoter. The retention of methylation in the PA-1 cell line could be due to the heterogeneous expression of the Oct4 protein in the cell population, with a decrease in the methylation frequency of one CpG island in the close proximity of the transcription start site, which could be critical for gene activation (arrow, ). In contrast, the p86-OTBC-L1 line was generated by serial selection and expansion of Oct4-positive cells, leading to an enrichment of Oct4 expression in the cell population. Consistently, this cell line exhibited a concomitant decrease in DNA methylation in multiple CpG islands along the promoter (arrows, ).
Designer ZFP containing a KRAB domain upregulate Oct4 in cell lines carrying a methylated promoter
To regulate the Oct4 gene promoter, we designed a 6ZFP recognizing an 18-base pair (bp) target located in the proximal promoter region at -119 base pairs upstream the translation start site of the gene. We chose this target sequence as it was found perfectly conserved in multiple vertebrate species, such as mouse and human, indicating an important regulatory function.Citation46 In addition, engineered ZFPs have been shown to efficiently regulate target genes in the close proximity of the translation start site (). The ZFs were constructed by a modular helix grafting method using the helical sequences known to interact with each of the corresponding triplets of targeted site ().
Figure 2. Design of an ATF/Artificial ZFP to upregulate the endogenous Oct4 promoter. (A) Schematic illustration of the Oct4 human promoter outlining the 18-bp ZF-119 targeted sequence and its location at position -119, relative to the translation start site (first-Met coding ATG triplet, +1). The ZF-119 sequence was chosen because of the high degree of conservation across vertebrate species, and because of its close proximity to the translation start site, which is typically accessible by TFs and nucleosome-free. Numbers designate the distance in bps relative to the start of translation. A putative binding site for SP1 is indicated in green; a putative Hormone Response Element (HRE) site is outlined with an open box; the Sox2 binding-site and its recognition sequence at position -236 are shown in blue; the Conservative Regions CR1 to CR4 within the promoter are shown in gray. Conservative Region 4 (CR4) contains the binding site for Oct4 and Sox2 proteins (red and blue circles and red and blue sequences, respectively). Arrows show the orientation of the 18-bp binding site in the promoter (from 5′ to 3′). (B) Schematic representation of the ZFPs generated in this study. The 6-Zinc Finger (ZF) arrays bound to DNA in absence of effector domains are indicated as ZF-119. Indicated below is an schematic representation of the different constructs, outlining the orientation of the effector domains linked to the ZF-119: KRAB, VP16, VP64 (4x VP16 domain) and Dnmt3a (C) Alphα-helical ZF amino acid sequences of ZF-119 engineered to bind their corresponding target DNA triplets (5′ to 3′). The helical residues at positions -1, +3 and +6 make specific contacts with the recognition triplets (blue refers to position -1, red for position +3 and purple for position +6).
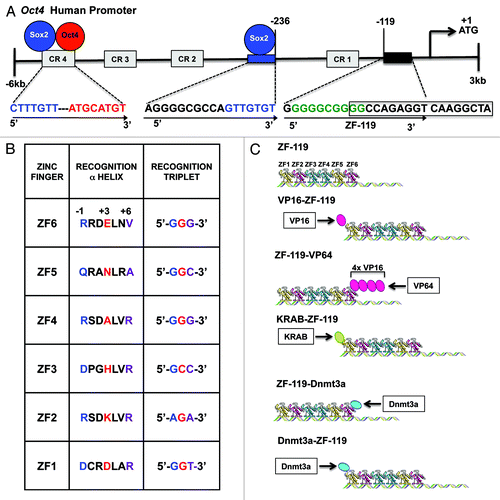
To test the capability of our engineered proteins to regulate the endogenous Oct4 target we linked the resulting 6ZF DNA-binding domain (ZF-119) with a panel of effector domains: 1. The N-terminal Herpes Virus 16 (VP16), which functions as transcriptional activator by recruiting the mediator protein. 2. The C-terminal VP64, which is a tetramer of VP16 engineered in the C-terminus of artificial ZFPs to upregulate target gene expression.Citation47 3. The Kruppel Associated Box (KRAB domain), which is located in the N-terminus of several natural ZF-containing proteins, and typically functions as a repressor domain.Citation48,Citation49 In addition, as controls, we took advantage of the catalytic domain of the DNA-methyltransferase 3a (Dnmt3a), which was positioned either in the N- or C-terminal end of ZF-119. In both configurations (N- and C-terminal fusions), the Dnmt3a catalytic domain has been shown to effectively silence targeted promoters when linked to 6ZF arrays.Citation38 In addition, we assessed the non-specific regulatory effects of the ZFP by cloning non-cognate 6ZF domains designed to bind other sequences of the genome (a non-specific 6ZF array and a KRAB-6ZF construct engineered to regulate the maspin promoter (ATF-126Citation31). The resulting fusions were cloned in a retroviral vector pMX-IRES-GFP and retroviral particles were used to infect different human cell lines carrying a silenced Oct4 promoter. An Oct4 cDNA expression vector was used as positive control for the transduction in the different host cell lines and real time expression analyses of all the panel constructs was normalized to hESCs. In the MDA-MB-231 cell line, the Oct4-specific 6ZFs were able to upregulate Oct4 only when linked to the KRAB domain (146.1 fold over control-transduced cells, which corresponds up to 15% of the amount of Oct4 mRNA expressed in hESCs). The ZF-119 (the DNA-binding domain in absence of effector function) and the ZF-119 linked to either a N-terminal or C-terminal activators were unable to activate the endogenous Oct4 (). In addition, both the N- and C-terminal ZF-119-Dnmt3a fusions did not significantly modulate Oct4 mRNA expression. The transduction of non-cognate (non-specific 6ZFs) or non-cognate 6ZFs linked to the KRAB domain was also un-effective in upregulating Oct4 (). The expression of several 6ZF-effector domain fusions was validated by western blotting and immunoprecipitation (Fig. S2 and ref. Citation32) and by immunofluorescence.Citation38 Overall, these results indicated that ZF-119 was able to upregulate Oct4 only in fusion with the KRAB domain. Moreover, the upregulation of Oct4 by the KRAB-ZF-119 construct was approximately one order of magnitude below the exogenous (ectopic) expression of the cDNA. In addition to Oct4, the KRAB-ZF-119 construct triggered a small but significant upregulation of the downstream target of Oct4 Nanog in MDA-MB-231 cells relative to control-transduced cells (10.7 fold).
Figure 3. KRAB-ZF-119 upregulates Oct4 expression in breast and ovarian cancer cell lines. (A-D) Quantification of Oct4, Sox2 and Nanog mRNA expression levels by qRT-PCR in MDA-MB-231 (A-B), SUM159 (C) and OVCAR-3 cells (D). X-axis represents the endogenous expression of these pluripotency TFs in the un-transfected cell line, together with cells transfected with the different ZFP designs. The Oct4 cDNA was transfected in the same pMX vector as positive control for the transfection; Y-axis represents the fold mRNA expression relative to hESCs. The ZFPs were retrovirally delivered in the cells and total mRNA was extracted for quantitative real-time PCR of gene expression and the results were normalized to hESC samples. Experiments were run in triplicate and data represent the mean ± SD of three independent experiments. Statistical significance between samples (e.g KRAB-ZF-119 vs. control) was analyzed by the Student’s t-test (***p < 0.0001; **p ≤ 0.01; *p ≤ 0.05). (E) Detection of Oct4 protein levels (upper panel) by western blot in OVCAR-3 cells transduced with either empty vector (control), VP16-ZF-119, Oct4 cDNA (positive control), KRAB-ZF-119, ZF-119 (no effector domain), ZF-119-VP64, or ZF-119-Dnmt3a. Non-transformed HMEC cells were used as negative control for Oct4 expression; NT2/D1 and p86-OTBC-L1 cells were used as positive cancer cell lines control expressing high levels of Oct4. An anti-Histone H3 antibody was used as loading control. The quantification of the bands of representative western blot is shown in (F). The Oct4 expression was normalized to the NT2/D1 sample. (G). Transduction of KRAB-ZF-119 and Oct4 cDNA in OVCAR-3 cells results in induction of tumorsphere formation. Representative images of the transfected cells are shown, outlining the morphological differences in the cells transfected with the KRAB-ZF-119 and Oct4 cDNA, which formed highly proliferative and compact spheroids. Control refers to empty-vector transduced cells. A quantification of the number of spheroids for each sample is shown in (H). Data representing three independent experiments was normalized to control-transduced cells. Statistical significance was analyzed by the Student’s t-test (***p < 0.0001; **p ≤ 0.01; *p ≤ 0.05).
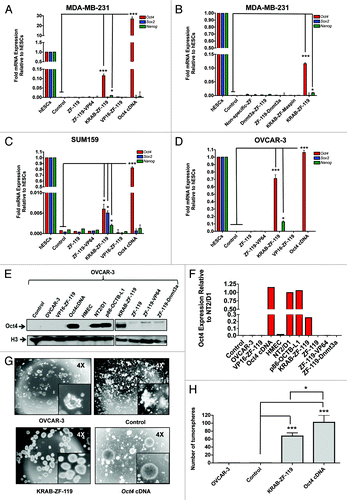
In the SUM159 cell line, the upregulation of Oct4 by KRAB-ZF-119 was 8 fold over control cells, which represents approximately 1% of the mRNA expression in hESCs (). The upregulation of Oct4 in this line was also accompanied by significant re-activation of Sox2, and in less extent Nanog mRNAs, which could be explained by the feed forward auto-regulatory feedback loop (transactivation) described between Oct4, Sox2 and Nanog TFs in ESCs.Citation6 Interestingly, in another silenced Oct4 cell line, OVCAR-3, the KRAB-ZF-119 upregulated both, Oct4 (951.1 fold relative to control, approximately 75% of the total mRNA detected in hESCs) and in less extent, Nanog (136.89 fold relative to control, representing up to 10% of the Nanog mRNA in hESCs; ). The lack of significant upregulation of Sox2 by KRAB-ZF-119 relative to control could be explained by the high levels of expression of this gene in this particular cell line (). The strong upregulation of Oct4 by KRAB-ZF-119 in the OVCAR-3 cell line was also validated by immunoblotting (). These data revealed that the endogenous expression levels of Oct4 in the KRAB-ZF-119 cells account for approximately 20% of the protein amount detected in NT2/D1 embryonic teratoma cells (), which exhibit Oct4 expression levels comparable to those in hESCs (). In summary, our gene expression analyses demonstrated that KRAB-ZF-119 was able to upregulate the endogenous Oct4 target in cancer cell lines carrying an epigenetically silenced promoter and that this re-activation of Oct4 was dependent on both, the KRAB effector function and an Oct4-specific (cognate) DNA-binding domain. To test whether more distally designed KRAB-ZF proteins could similarly upregulate the Oct4 promoter, we designed a second protein binding an 18-bp site located 395-bp upstream the translation start site. This more distally designed ZF protein was unable to reactivate Oct4 when attached to a KRAB or other effector domains (Fig. S3). These results outline the importance of the precise positioning of the engineered ZF-binding site in the endogenous proximal promoter for effective regulation of Oct4.
The phenotypical consequences of Oct4 upregulation were consistent with previously published reports, which suggest that Oct4 increases the expansion of a poorly differentiated stem cell-like population.Citation12,Citation50 As expected, both Oct4 cDNA and in less extent KRAB-ZF-119, were able to dramatically induce the formation of compact self-renewal spheroids or tumorspheres in the OVCAR-3 cell line, over control-transduced cells ( and H). Tumorsphere formation assays monitor the capability of single cells to proliferate in low adherence plates, and is an in vitro assessment of stemness and tumorigenic potential.
Subsequently, in vivo chromatin immunoprecipitation (ChIP) experiments were performed to physically demonstrate a direct association between the ZF-119 and its cognate site in the Oct4 promoter. The ZF-119 expression construct was engineered with a C-terminal Human influenza hemagglutinin (HA) tag, which allowed the immunoprecipitation of the ZF-119-DNA complexes in transduced cells. We chose the SUM159 line in these experiments since these cells were highly proliferative and permitted the collection of high number of cells (> 105 cells) required for ChIP experiments. The ChIP elutes from either control or ZF-119-transduced cells were amplified using specific primers flanking the ZF-119 binding site. As shown in , a specific ChIP product was amplified in cells transduced with ZF-119 but not in control-transduced cells, indicating that ZF-119 was binding its cognate site in the Oct4 promoter.
Figure 4. KRAB-ZF-119 binds the endogenous Oct4 promoter and upregulates transcription. (A) Chromatin immunoprecipitation (ChIP) to detect the binding of ZF-119 to its target site in SUM159 cells. An anti-HA antibody was used to capture the ZF-119 bound to its genomic target DNA from control-transduced cancer cells and cells transduced with ZF-119. A quantification of the ChIP assay by densitometric analyses of the bands from the same gel is outlined below. Data was normalized to the input signal. (B) Schematic representation of a proposed transcriptional activation model in the Oct4 gene promoter mediated by KRAB domain. After KRAB-ZF-119 binds to the Oct4 gene promoter, the KRAB domain could recruit and/or facilitate the recruitment of a constellation of transcriptional activating factors including Sox2, facilitating their access and binding to the Oct4 promoter; alternatively the KRAB or KRAB-associated factors could inhibit a transcriptional repressor complex in the proximal promoter resulting in transcriptional activation of the gene promoter.
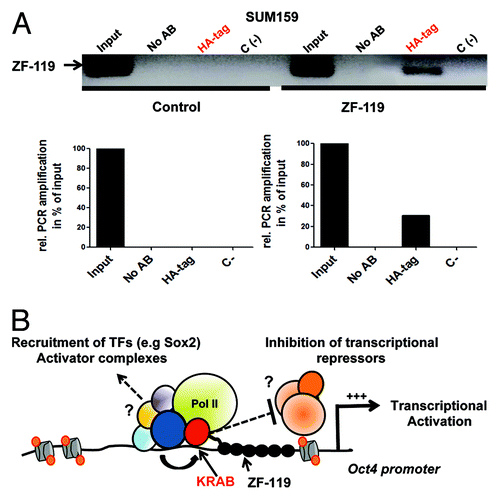
Putative mechanisms by which the ZF arrays mediate the DNA-binding and access to chromatin in the context of a silenced Oct4 are outlined in . An N-terminal KRAB domain could mediate Oct4 upregulation the activation of Oct4 by facilitating the binding of TFs (e.g., Sox2), co-factors and chromatin remodelers, which are necessary for transcriptional activation. KRAB-ZF-119 could either indirectly (e.g., by changing chromatin structure) or by physically interacting with its associated factors in the promoter. Another possible mechanism of action, would involve a negative effect of KRAB-ZF-119 on repressive epigenetic regulators, such as Dnmt3a or Dnmt3b, or other transcriptional repressors by KRAB or KRAB-associated proteins, a model that has also been proposed in the context of natural KRAB-containing ZFs.Citation39,Citation40 It is also important to remark that the ZF-119 binding site overlaps with a SP1 site in the 5′ of the 18-bp duplex, and with a putative Hormone response Element (HRE) site in the 3′ of the binding site. Thus, it is also possible that KRAB-ZF-119 could negatively affect the binding and transcriptional activity of SP1 and/or HR or other associated factors in the context of the Oct4 promoter.
Discussion
Oct4 encodes a homeodomain-containing TF, which is essential to induce and maintain a pluripotent ES or ES-like state.Citation1 While Oct4 is expressed at high levels in ESCs, its promoter undergoes several layers of epigenetic silencing in differentiated cells.Citation44 In this paper we investigated the capability of engineered ZFPs to upregulate the endogenous Oct4 gene in cell lines carrying an epigenetically silenced promoter.
First, we examined the expression levels of pluripotency TFs Oct4, Sox2 and Nanog in a panel of breast and ovarian cancer cell lines. We found that Oct4 and Nanog were transcriptionally silenced in the vast majority of the breast cancer cell lines examined. Sox2 mRNA was expressed in some breast cancer cell lines, such as MCF-7 and ZR-75–1. In contrast, the pluripotency TFs were abundantly expressed in ovarian cancer cell lines and ovarian cancer cell lines derived from serous ovarian cancer (SOC) patients. In a recent report, Oct4 expression was found enriched in ascites from ovarian cancer patients that underwent resistance to chemotherapy.Citation13 The tissue localization of the Oct4 protein in ovarian cancer cell lines and tumor specimens was investigated by IF and IHC, respectively. We found that Oct4 was predominantly nuclear while some cytoplasmic staining was also detected, particularly in tumor specimens, in accordance with other recent studies.Citation14 The significance of the cytoplasmic staining of Oct4 and other homeodomain proteins is not well understood. However, protein-protein interactions between homeodomain-containing TFs and translation initiation factors have been described outlining a possible action of these TFs regulating translation and/or mRNA transport.Citation51 The lack of expression of Oct4 in breast and some ovarian cancer cell lines was associated with DNA methylation. Moreover, cell lines expressing Oct4, such as PA-1 line, also retained DNA methylation frequencies in the promoter. This is in contrast with fully reprogrammed cells and ESCs, which have an Oct4 promoter free of DNA methylation.Citation15,Citation16,Citation18,Citation45 In gene promoters, DNA methylation positively correlates with H3K9me and cross-talks mechanisms between these marks have been reported via direct interactions between DNA methyltransferases and Histone methyltransferase G9a.Citation42,Citation52 Thus, additional layers of epigenetic silencing mechanisms are anticipated to act in conjunction with DNA methylation in the Oct4 promoter context.
Reactivation of Oct4 in breast cancer cells has been associated with oncogenesis and tumor initiation.Citation12 Ectopic delivery of Oct4 cDNA in breast epithelial cells resulted in the reactivation of the endogenous gene, which triggered the formation of highly proliferative spheroids or tumorspheres in low adherence plates. These Oct4+ cells were able to generate tumors in xenograft models of breast cancer and as few as 50 cells gave rise to poorly differentiated, aggressive tumors presenting stem cell-like features and colonization capabilities.Citation12 Reciprocally, siRNA-mediated knock-down of Oct4 and Oct4 targets, such as Nanog, dramatically abolished the formation of tumorspheres in vitroCitation12 and induced apoptosis of breast cancer cells.Citation53 Likewise, high Oct4 expression has been reported in aggressive epithelial ovarian cancers and ascites, and has been associated with tumor initiation, chemoresistance, unfavorable prognosis and disease recurrence.Citation13,Citation14,Citation54-Citation58 Our results are consistent with these functions of Oct4 in tumor initiation and carcinogenesis. As expected, endogenous Oct4 upregulation by KRAB-ZF-119 in OVCAR-3 cells dramatically induced the formation of compact tumorspheres in non-adherent plates, suggesting increased tumorigenic potential.
More recently, Oct4 expression has been detected in breast cancers associated with pregnancy and lactation (Hassiotou et al., submitted manuscript). These pregnancy-associated breast cancers often lack the expression of cellular receptors (ER, PR, ErbB2) and are associated with poor clinical outcome.Citation59,Citation60 The epigenetic silencing mechanisms both operating at DNA and at histone level could cooperate to ensure that in somatic cells, Oct4 is maintained in a repressed state, protecting the cell from oncogenic activation and tumor initiation. Our data suggest that some aggressive cancers, possibly enriched in tumor initiating cells, express Oct4. Re-activation of Oct4 could occur from more differentiated epithelial cells by a partial loss of DNA methylation and possibly in conjunction with other histone post-transcriptional modifications. Alternatively, a pre-existing Oct4+ cancer-stem-cell like population could give rise to more differentiated cells acquiring de novo DNA methylation and gene silencing. In the vast majority of the tumor samples analyzed, no overall correlation between Oct4, Sox2 and Nanog mRNA expression levels was observed. This is in contrast with ESCs where a positive feed forward loop between these TFs has been proposed, resulting in high expression of all three TFs. Our results suggest that aberrant epigenetic reactivation of either Oct4, Sox2 or Nanog could independently occur in the cancer cells to drive oncogenic activation.
Subsequently, we used ZF technology to address the capability of artificial DNA binding proteins to re-activate a silenced Oct4 promoter in cell lines carrying high levels of DNA methylation. We constructed an artificial 6ZF protein targeting an 18-base pair (bp) site located 119 bps upstream the translation start site. Each ZF is a quasi-modular unit that can be engineered to recognize specific recognition triplets. The ZF-119 site was chosen since it was conserved among several vertebrate species, possibly reflecting an important function for the regulation of the promoter. Indeed, the ZF-119 targeted site partially overlapped with a putative SP1 binding site (5′-GGGGCGGGG-3′) in the 5′ of the 18-bp duplex, and with a Hormone Response Element (HRE, 5′-GGCCAGAGGTCAAGGCTA-3′) site in the 3′ of the duplex. The flanking sequences immediately upstream and downstream the ZF-119 site contained highly conserved G/G rich motifs.Citation46 In addition, a Sox2-binding site was located in the proximity (~117 bp) of the ZF-119 binding site.
Previously, we have described artificial ZFPs or Artificial Transcription Factors (ATFs) able to bind and reactivate the tumor suppressor mammary serine protease inhibitor (maspin) in lung and breast cancer cell lines carrying an epigenetically silenced promoter. The ATFs reactivated maspin only when the 6ZF DNA-binding domains were linked to an activator domain (VP64).Citation30-Citation32 The 6ZF-VP64 arrays demonstrated high potency and specificity in upregulating maspin, with expression levels similar to exogenous maspin cDNA delivery.Citation33
In contrast with maspin, ZF-119 only reactivated Oct4 when the 6ZFs where linked to a Kruppel Associated Box (KRAB) domain. This result was not initially anticipated since the KRAB domain has historically been regarded as transcriptional repressor.Citation48 Importantly, the potency of the re-activation by KRAB-ZF-119 was dependent on the cell line analyzed. The highest regulatory potential of the protein was found in the OVCAR-3 ovarian cancer cell line, where KRAB-ZF-119 was able to upregulate the endogenous Oct4 with mRNA levels approaching 75% of those detected in hESCs. This cell line-dependency, in despite of the high levels of DNA methylation and gene silencing, suggest that the chromatin microenvironment is different in each line, possibly influencing the regulatory potential of the ZFP. For example, one difference between OVCAR-3 and MDA-MB-231 cells is the very high levels of Sox2 in OVCAR-3, which could prime or facilitate an activation state in the promoter. Finally, in the vast majority of cell lines analyzed re-activation of Oct4 by KRAB-ZF-119 was also accompanied by a significant upregulation of the Oct4 downstream targets, such as Sox2 and Nanog, as expected from the auto-regulatory feedback-loop previously described between Oct4-Sox2-Nanog in ESCs.Citation6 However, we cannot exclude that KRAB-ZF-119 could also be recruited to other promoters and thus ChIP-Seq data will be necessary to map all ZFP binding sites in the cancer cell genome.
The KRAB domain is particularly abundant in natural ZF TFs hence it has been mapped in the N-terminus of the DNA-binding domain in about one third of all C2H2 ZFPs. The KRAB domain spans ~75 amino acids present exclusively in vertebrates, and is composed of two boxes, A and B. One of the main co-repressors of KRAB is KRAB-Associated protein 1 (KAP-1), which interacts with box A.Citation49 In the proposed model of repression, KAP-1 can recruit the chromatin modifier SETDB1, which directs H3K9me3.Citation61 In addition, KAP1 also binds Heterochromatin protein 1 (HP1), which interacts with H3K9me3, stabilizing the repressive complex in chromatin. Recent genome-wide studies revealed that most KRAB-ZFPs recruit KAP-1 to the 3′-coding exons of ZF genes, and not to the gene promoters. It has been proposed that KAP-1 could be recruited to promoter sequences through interaction with other DNA-binding TFs.Citation39,Citation40
Although the function of KRAB as a repressor has been long described, recent genome-wide data from ChIP-Seq had demonstrated that natural KRAB-ZFPs bind and activate certain cellular targets. One example is the ZFP236, a TF that contains a N-terminal KRAB domain. While ZFP263 acts as a repressor for some cellular targets, the same protein binds and upregulates other sets of cellular promoters, such as foxa1, itpka and rgs10.Citation41 These data suggest that although ZFP containing KRAB domains function as transcriptional repressors, in some genomic contexts they can also act as activators. Our results are in agreement with this new emerging function of KRAB-containing ZF proteins in gene activation. In addition, the reprogramming field has recently reported numerous examples ZFPs, including N-terminal KRAB or other repressor domains (such as SCAN domains) that participate in the upregulation of self-renewal gene promoters. One example is ZFP206 (Zscan10), a TF expressed in ES cells and an integral component of the self-renewal circuitry. The overexpression of ZFP206 activated transcription of some direct targets such as Oct4, Klf5 and Jarid1c, while repressing the expression of others promoters, such as Meis2, mir-124-a and mir-124-a2.Citation62 These results reinforce the idea that genomic context can affect the specificity and the regulatory potential of ZFPs. As with KRAB-domains, SCAN domains could also mediate context-dependent protein-protein interactions at genomic targets, resulting in either activation or repression. The particular composition of co-activators, co-repressors and epigenetic modifiers in targeted regulatory regions is potentially critical for the recruitment of ZFPs and for the regulatory outcome of these proteins.
The mechanism of action of KRAB-containing ZFPs as transcriptional activators still remains elusive and will require further investigation. KRAB-associated proteins could negatively interfere with repressive proteins, such as Histone deacetylases, Histone methyltransferases, DNA methyltransferase and other complexes. The ZF-119-binding site overlaps with a putative SP1 binding site and with a HRE site, and it is possible that the engineered protein could negatively affect the transcriptional activity of these endogenous proteins. Alternatively, KRAB-ZFPs may also function to facilitate the binding of TFs and co-activators into the targeted promoter. KRAB-ZF-119 could be regarded as a “pioneer transcription factorCitation63”: by binding into the proximal Oct4 promoter, KRAB-ZF-119 could directly (e.g., via direct recruitment) or indirectly (e.g., via local changes in chromatin structure) engage the access of Sox2 and/or other TFs in the Oct4 promoter. Several ZFPs involved in the maintenance of a pluripotent state, such as ZFP206Citation41 and ZFP281,Citation64 have been shown to physically interact with self-renewal TFs, such as Oct4, Sox2 and Nanog. Similarly, KRAB-ZF-119 could physically interact with KRAB-associated factors and recruit TFs, co-activators and chromatin remodelers to activate targeted genes. For example, KRAB-ZF-119 could recruit self-renewal TFs, such as Sox2, which is abundantly expressed in SOC cell lines, such as OVCAR-3 and PA-1. Further research is required to validate these models and to elucidate the exact mechanism of action of KRAB-ZFPs. Another interesting question for further study is whether TAL Effectors containing N-terminal KRAB domains are also subjected to the same regulatory outcomes as ZF-based ATFs. One difference between ZF proteins and TALE effectors is the size of the DNA binding domains, ZFs being more compact and particularly biased to recognize GC-rich sequences, which are abundant in proximal promoters. In conclusion, our data supports a new paradigm for KRAB-ZFPs and suggests that these proteins could function, in certain genomic loci, as activators of endogenous genes. These findings have important implications for investigators working within the areas of ZF protein biology, genomic reprogramming, artificial gene regulation and cell engineering.
Materials and Methods
ATF construction
Target sites for ZF recognition within the Oct4 human proximal promoter (GenBank accession number AJ297527.1) were selected using the website www.zincfingertools.org.Citation65 ZF-119 was selected based on its close proximity to the translation start site and because the 18-bp site was perfectly conserved across vertebrate species, including human and mouse.Citation46 Specific primers were designed to incorporate the amino acid residues in the ZF’s recognition helix of ZF-119, responsible for the binding to the target sequence. The 18-bp target site of ZF-119, shown in , is: 3′GGGGGCGGGGCCAGAGGT-5′ while the Oct4-ZF sequences are shown in . The ZF-119 DNA-binding domain was generated by overlapping PCR as previously described.Citation31 The PCR fragments were digested with SfiI and sub-cloned into the retroviral vector pMX-IRES-GFP containing different effector domains: a N-terminal VP16, a C-terminal VP64,Citation31 a N-terminal Dnmt3a,Citation38 a C-terminal Dnmt3aCitation38 and a N-terminal KRAB,Citation37 producing the following ATFs: pMX-VP16-ZF119, pMX-ZF119-VP64, pMX-Dnmt3a-ZF119, pMX-ZF119-Dnmt3a and pMX-KRAB-ZF119. Each ATF contains an internal SV40 nuclear localization signal (NLS) and a terminal hemagglutinin (HA) decapeptide tag. Plasmid sequencing was performed to confirm the correct nucleotide sequence of each ATF construct.
Cell lines and cell culture
Human breast cancer cell lines SUM102, SUM149, SUM159, MCF7, MCF12A, MDA-MB-231, MDA-MB-435s, MDA-MB-453, MDA-MB-468, ZR-75–1 and BT-543 were obtained from the Core Tissue Culture Facility of the UNC Lineberger Comprehensive Cancer Center (Chapel Hill, NC). The cell line p86-OTBC-L1 was isolated previously by our group.Citation12 The packaging cell line 293TGagPol and the human testicular embryonic carcinoma cell line NT2/D1 were obtained from the American Type Culture Collection (ATCC). Cell lines were propagated in growth medium as specified by the ATCC and maintained at 37°C and 5% CO2. The packaging 293 T-GagPol cells, NT2/D1 and the human breast cancer cell lines MDA-MB-231, MDA-MB-435s, MDA-MB-453 and MDA-MB-468 were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 10% Fetal Bovine Serum (FBS, BenchMark, Gemini Bio Products, Sacramento, CA) and 1% Penicillin/Streptomycin (Pen/Strep, Invitrogen, Carlsbad, CA). Culture media of MDA-MB-435s cells contained additionally 0.01 mg/ml Bovine Insulin (Gibco/ Invitrogen, Carlsbad, CA). MCF7 breast cancer cells were cultured in Minimum Essential Media (MEM) supplemented with 1.5 g/l sodium bicarbonate, 0.1mM non-essential amino acids (NEAAs), 1mM sodium pyruvate, 0.01 mg/ml Bovine Insulin, 10% FBS and 1% Pen/Strep. MCF7 cells were cultured in DMEM containing 20 ng/ml Epithelial Growth Factor (EGF), 100 ng/ml cholera toxin, 00.1 mg/ml Bovine Insulin, 500 ng/ml hydrocortisone, 5% Horse serum and 1% Pen/Strep. MDA-MB-468 breast cancer cells were cultured in L15 media supplemented with 10% FBS and 1% Pen/Strep. ZR-75–1 and BT549 cells were cultured in RPMI 1640 media supplemented with 10% FBS and 1% Pen/Strep. SUM102 and SUM149 cells were cultured in human mammary epithelial cell (HMEC) media containing HMEC supplemental bullet kit (Gibco/Invitrogen Carlsbad, CA), bovine pituitary extract (Gibco/Invitrogen) and 1% Pen/Strep. For SUM149 cells, the media contained additionally 5% FBS. SUM159 breast cancer cells were cultured in Ham’s F12 media containing 5 mg/ml Bovine Insulin, 1 mg/ ml hydrocortisone, 10mM Hepes, 5% FBS and 1% Pen/Strep. SK-Br-3 cells were cultured in McCoy’s 5a Medium, supplemented with 10% FBS and 1% Pen/Strep. MDA-MB-453s breast cancer cells were cultured in Leibovitz’s L15 Medium, supplemented with 10% FBS and 1% Pen/Strep. Human ovarian cancer cell line OVCAR-3 were culture in RPMI medium supplemented with 20% FBS and 1% Pen/Strep. Human ovarian teratocarcinoma PA-1 cells were cultured in MEM α medium supplemented with 10% FBS and 1% Pen/Strep. Human epithelial adenocarcinoma SKOV-3 cells were cultured in McCoy medium supplemented with 10% FBS and 1% Pen/Strep.
Ovarian cancer patient-derived samples
Ascites were obtained from the Pontificia Universidad Católica de Chile with written patient consent and approved IRB protocols. Ascites cells were grown in adherent tissue culture plates in DMEM/F12 medium (Gibco/Invitrogen, Carlsbad, CA), containing 1% antibiotic/antimitotic (Gibco/Invitrogen, Carlsbad, CA), 5% FBS (BenchMark, Gemini Bio Products, Sacramento, CA).
RNA and DNA preparation
Cell cultures were harvested for RNA and DNA preparation before and after retroviral transduction. To purify total RNA, cell samples derived from patients were resuspended in Trizol reagent following the manufacturer’s instructions (Gibco/Invitrogen, Carlsbad, CA). Cell line culture samples were processed with the RNeasy kit (Qiagen, Valencia, CA) and processed for qRT-PCR analyses as described below. A Puregene DNA Purification Kit (Gentra Systems, Minneapolis, PA) was used to purify DNA while genomic DNA was isolated using the DNeasy Tissue Kit (Qiagen, Valencia, CA).
Sodium bisulfite conversion and DNA methylation mapping in the Oct4 promoter by Sequenom EpiTYPER assay
The methylation status of 19 CpG dinucleotides within the human Oct4 gene promoter region was examined in bisulfide-modified DNA as previously described.Citation38 Genomic DNA (1.5 μg) isolated from un-transduced cell line samples (Fibroblast, HMEC, MDA-MB-231, OVCAR-3, PA-1 and p86-OTBC-L1) was modified using the EZ DNA Methylation-Gold Kit (Zymo Research, Irvine, CA). Samples were further purified with the Wizard DNA purification resin (Promega, Alexandria, NSW) and resuspended in water. Bisulfite converted DNA was processed using a Sequenom EpiTYPER platform at the Yale Center for Genome Analysis (Yale University, West Haven, CT). The primers used for the amplification of converted DNA were: Oct4-amp1-F (forward primer): TAGTTGGGATGTGTAGAGTTTGAGA; Oct4-amp1-R (reverse primer): TAAACCAAAACAATCCTTCTACTCC; Oct4-amp8-F: GGAGTAGAAGGATTGTTTTGGTTTA; Oct4-amp8-R: ATCACCTCCACCACCTAAAAAA; Oct4-amp14-F: TTTATTATTTTTATTATTTGGAGGGGG; Oct4-amp14-R: CAATAATCCAAACCTATAATCCCAA.
ATF retroviral transduction
The retroviral vector pMX-IRES-GFP containing the Oct4 ATFs (pMX-ZF119-VP16, pMX-ZF119-VP64, pMX-ZF119-Dnmt3a, pMX-Dnmt3a-ZF119 or pMX-ZF119-KRAB) was first co-transfected with the pMDG.1 plasmid, expressing the vesicular stomatitis virus envelope protein into the packing 293TGag-Pol cells to produce retroviral particles. Transfection was performed using Lipofectamine according with the manufacturer’s instructions (Gibco/Invitrogen, Carlsbad, CA). The viral supernatant was collected and used to infect the host cell lines. Transfected cells were incubated at 37°C in 5% CO2 for 4 d, and collected for RNA, DNA or protein extracts. The RNA was extracted, reverse transcribed and processed for real-time quantification of Oct4 mRNA expression.
Quantitative real-time PCR
RNA from cell line cultures and patient-derived samples was extracted as mentioned above according to the manufacturer’s recommendations and reverse-transcribed to cDNA using the High Capacity cDNA reverse transcription kit (Applied Biosystems, Carlsbad, CA). For qRT-PCR assays, reactions were performed with a final concentration of 1 × of TaqMan Fast Universal Master Mix (Applied Biosystems, Carlsbad, CA) and 150 ng of cDNA. Primers used for qRT-PCR assays are described by Beltran et al.Citation12 qRT-PCR data was analyzed with the 7500 Software Version 2.0.5, using the comparative ΔCt method and GAPDH as an endogenous reporter gene (AB Prism software, Applied Biosystems; Foster City, CA). The results were plotted as the average of at least 3 independent experiments with duplicates, and standard errors calculated using GraphPad Prism v5 software. Fold-Oct4 mRNA levels were calculated relative to the fibroblast sample or, for transduced experiments, to the control cells (cells transduced with an empty retroviral vector, when indicated). Statistical differences between groups were determined by Student’s t-test.
Western blotting
Total protein extract was obtained with Radioimmunoprecipitation Assay Buffer (RIPA buffer; Sigma-Aldrich, St Louis, MO). Nuclear protein extraction was performed using the NE-PER Nuclear and Cytoplasmic Extraction Kit (Pierce Biotechnology, Rockford, IL). Protein lysates (50 µg) were resolved on 12% SDS-PAGE gels. Antibodies included: rabbit anti-Oct4 (Abcam, Cambridge, MA) diluted at 1:250; anti-Tubulin (Sigma-Aldrich, St Louis, MO) diluted 1:5000; anti-H3 (Active Motif, Carlsbad, CA) diluted 1:10,000, and horseradish peroxidase-conjugated antibody, sheep anti-mouse and donkey anti-rabbit (1:5000, GE Healthcare, Piscataway, NJ). Western blotting detection was performed with the Amersham ECL Detection System (GE Healthcare, Waukesha, WI). Western blots were quantitated using the program Image J v1.46 (ImageJ, NCBI, Bethesda, MD). Expression of Oct4 protein in each cell preparation was normalized to the NT2/D1 cell line.
Immunoprecipitation of the ZF fusions
MDA-MB-231 cells were retrovirally transfected as previously described. Forty-eight hours post-transfection, cells were collected and total protein extract isolated with Radioimmunoprecipitation Assay Buffer (RIPA buffer; Sigma-Aldrich, St Louis, MO). Protein lysates (500 µg) were pre-incubated with 1 μg of anti-HA antibody (Covance, Indianapolis, IN) for 2h at 4°C and 40 µl of protein A/protein G beads (Santa Cruz, CA) were added to each sample and incubated in constant agitation for 24h. Samples were collected by centrifugation at 3000 rpm (~1000 xg) for 30 sec at 4°C. Supernatant was discarded and pellet was gently washed 4 times with 1ml of RIPA buffer, followed of 4 rinse steps with 1 × PBS buffer. After the final rinse, beads were resuspended in 40 μl of 2 × electrophoresis sample buffer (Santa Cruz 24945) and resolved on 12% SDS-PAGE gel. Immunodetection was performed with an anti-HA antibody (Covance, Indianapolis, IN).
Chromatin immunoprecipitation (ChIP)
ChIP assay was performed to detect the ZF-119 binding to the human Oct4 gene promoter in vivo. Briefly, ~107 SUM159 cells were transduced with either control (empty retroviral vector) or ZF-116. Protein-DNA complexes were pulled-down with protein A/protein G beads (Santa Cruz, CA) coupled with anti-HA antibody (Covance, Indianapolis, IN). After incubation, the beads were washed out three times with low-salt buffer, three times with high-salt buffer and two times with Tris-EDTA buffer. The DNA was eluted from the beads by incubating them overnight at 65°C with an elution buffer (Tris-EDTA buffer, 1% SDS and 2 μl of Proteinase K). DNA was purified by phenol/chloroform-extraction procedure. The immunoprecipitated DNA was amplified by PCR, using the following primers flanking the ZF-119 binding site in the Oct4 gene promoter: 5′-ACCATTAGGCAAACATCCTT-3′ (forward primer) and 5′-GGGGAAGGAAGGCGCCCC-3′ (reverse primer). As control for PCR amplifications the house-keeping gene GAPDH was used with the following primers: 5′-TACTCGCGGCTTTACGGG-3′ and 5′-TGGAACAGGGAGGAGCAGAGAGCA-3′. PCR products were visualized by agarose gel electrophoresis and the relative fold enrichment determined by densitometry (ImageJ, NCBI, Bethesda,MD) and normalized to the input samples.
Immunofluorescence
Fibroblast, HMEC, MDA-MB-231, OVCAR-3, PA-1 and p86-OTBC-L1 cells were seeded in fibronectin-coated coverslips and fixed with 4% formaldehyde-PBS solution at room temperature for 10 min. Cells were permeabilized with 0.3% Triton X/PBS for 15 min at 4°C and 5% BSA/PBS was added for overnight blocking at 4°C. Fibronectin, formaldehyde and Triton-X were purchased from Sigma (St. Louis, MO). Cells were incubated at 4°C overnight with primary rabbit anti-Oct4 antibody (Abcam, Cambridge, MA) diluted 1:250 in 1% BSA/PBS. After the incubation, the slides were rinsed and labeled with the secondary antibody Alexa 555 anti-rabbit IgG in a 1:1,000 dilution (Gibco/Invitrogen, Carlsbad, CA). Nuclear staining was visualized with DAPI or Hoechst dye (Gibco/Invitrogen, Carlsbad, CA). Intracellular expression of Oct4 was detected using a Zeiss 510 Meta Inverted Laser Scanning Confocal Microscope at the Michael Hooker Microscopy Facility at UNC.
Immunohistochemistry (IHC)/Chromogenic detection of Oct4 in breast/ovarian cancer specimens
IHC was performed in the Bond Autostainer (Leica Microsystems Inc. Norwell MA 02061). Briefly, slides were dewaxed in Bond Dewax solution (AR9222) and hydrated in Bond Wash solution (AR9590). Antigen retrieval was performed for 30 min at 100°C in Bond-Epitope Retrieval solution 1 pH-6.0 (AR9961) and blocked with Dako Serum-Free Ready-To-Use Protein Block for 10 min (X0909). Slides were incubated with an anti-Oct4 antibody (ab18976 Abcam, Cambridge, MA) for 2 h. Antibody detection was performed using the Bond Polymer Refine Detection System (DS9800). Stained slides were dehydrated and coverslipped. Stained slides were imaged using the Aperio Scanscope XT (Aperio Technologies, Vista, CA 92081). Images were examined using Spectrum digital pathology platform.
Tumorsphere assay
OVCAR-3 cells were transfected with control, KRAB-ZF-119 or Oct4 cDNA retroviral vectors as described above. Seventy-two hours post-transfection 20,000 cells were seeded in triplicate wells, in a 6-well plate (low adherent plates, Corning, NY) in spheroid media: HMEC medium (Gibco/Invitrogen, Carlsbad, CA) containing 20 ng/ml human EGF (BD Biosciences), 1 μg/ml hydrocortisone (Stem Cell Technologies, Vancouver, Canada), insulin 5 μg/ml (Sigma) and 1 × B27 (Gibco/Invitrogen, Carlsbad, CA). The plates were incubated at 37°C in 5% CO2. Tumorspheres were visualized using a cell culture Leica microscope 3 d after seeding. Tumorspheres were counted and the mean ± SD of three independent experiments was plotted and normalized to OVCAR-3 cells transfected with an empty vector (control). Statistical differences between groups were determined by Student’s t-test (***p < 0.0001; **p ≤ 0.01; *p ≤ 0.05).
| Abbreviations: | ||
| ATFs | = | artificial transcription factors |
| DBD | = | DNA-binding domain |
| ZF | = | zinc finger |
| ZFPs | = | ZF proteins |
| TF | = | transcription factor |
| SKD | = | Super KRAB domain |
| KRAB | = | Kruppel Associated Box |
| BSA | = | Bovine Serum Albumin |
| NLS | = | nuclear localization signal |
| HA | = | terminal hemagglutinin |
Additional material
Download Zip (3.8 MB)Acknowledgments
We thank Michelle Mathews at the UNC Anatomic Pathology Translational Core Laboratory (APTCL) for expert technical assistance, and to Irina Tikhonova at the Yale Center for Genome Analysis (YCGA) for the assistance and analysis of DNA methylation. The UNC APTCL is supported, in part, by grants from the UNC University Cancer Research Fund (UCRF). This work was supported by National Cancer Institute/National Institutes of Health grants 1R01CA125273, 3R01CA125273–03S1, the Department of Defense (DoD) W81XWH-10–1-0265 (P.B.), the Chilean grants BMRC CTU06 (G.O.) and FONDECYT 1120292 (M.C.), the Chilean government science and technology grant CONICYT 21100327 (R.E.), and the Researcher Professor Grant by the Fulbright García-Robles Foundation (COMEXUS) (K.J.M.).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Sterneckert J, Höing S, Schöler HR. Concise review: Oct4 and more: the reprogramming expressway. Stem Cells 2012; 30:15 - 21; http://dx.doi.org/10.1002/stem.765; PMID: 22009686
- Walker E, Ohishi M, Davey RE, Zhang W, Cassar PA, Tanaka TS, et al. Prediction and testing of novel transcriptional networks regulating embryonic stem cell self-renewal and commitment. Cell Stem Cell 2007; 1:71 - 86; http://dx.doi.org/10.1016/j.stem.2007.04.002; PMID: 18371337
- Liang J, Wan M, Zhang Y, Gu P, Xin H, Jung SY, et al. Nanog and Oct4 associate with unique transcriptional repression complexes in embryonic stem cells. Nat Cell Biol 2008; 10:731 - 9; http://dx.doi.org/10.1038/ncb1736; PMID: 18454139
- Pardo M, Lang B, Yu L, Prosser H, Bradley A, Babu MM, et al. An expanded Oct4 interaction network: implications for stem cell biology, development, and disease. Cell Stem Cell 2010; 6:382 - 95; http://dx.doi.org/10.1016/j.stem.2010.03.004; PMID: 20362542
- Wang J, Rao S, Chu J, Shen X, Levasseur DN, Theunissen TW, et al. A protein interaction network for pluripotency of embryonic stem cells. Nature 2006; 444:364 - 8; http://dx.doi.org/10.1038/nature05284; PMID: 17093407
- Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell 2005; 122:947 - 56; http://dx.doi.org/10.1016/j.cell.2005.08.020; PMID: 16153702
- Babaie Y, Herwig R, Greber B, Brink TC, Wruck W, Groth D, et al. Analysis of Oct4-dependent transcriptional networks regulating self-renewal and pluripotency in human embryonic stem cells. Stem Cells 2007; 25:500 - 10; http://dx.doi.org/10.1634/stemcells.2006-0426; PMID: 17068183
- Feldman N, Gerson A, Fang J, Li E, Zhang Y, Shinkai Y, et al. G9a-mediated irreversible epigenetic inactivation of Oct-3/4 during early embryogenesis. Nat Cell Biol 2006; 8:188 - 94; http://dx.doi.org/10.1038/ncb1353; PMID: 16415856
- Gu P, Xu X, Le Menuet D, Chung AC, Cooney AJ. Differential recruitment of methyl CpG-binding domain factors and DNA methyltransferases by the orphan receptor germ cell nuclear factor initiates the repression and silencing of Oct4. Stem Cells 2011; 29:1041 - 51; http://dx.doi.org/10.1002/stem.652; PMID: 21608077
- Dyce PW, Toms D, Li J. Stem cells and germ cells: microRNA and gene expression signatures. Histol Histopathol 2010; 25:505 - 13; PMID: 20183803
- Hassiotou F, Beltran A, Chetwynd E, Stuebe AM, Twigger AJ, Metzger P, et al. Breastmilk is a novel source of stem cells with multilineage differentiation potential. Stem Cells 2012; 30:2164 - 74; http://dx.doi.org/10.1002/stem.1188; PMID: 22865647
- Beltran AS, Rivenbark AG, Richardson BT, Yuan X, Quian H, Hunt JP, et al. Generation of tumor-initiating cells by exogenous delivery of OCT4 transcription factor. Breast Cancer Res 2011; 13:R94; http://dx.doi.org/10.1186/bcr3019; PMID: 21952072
- Latifi A, Luwor RB, Bilandzic M, Nazaretian S, Stenvers K, Pyman J, et al. Isolation and characterization of tumor cells from the ascites of ovarian cancer patients: molecular phenotype of chemoresistant ovarian tumors. PLoS One 2012; 7:e46858; http://dx.doi.org/10.1371/journal.pone.0046858; PMID: 23056490
- Zhang J, Li YL, Zhou CY, Hu YT, Chen HZ. Expression of octamer-4 in serous and mucinous ovarian carcinoma. J Clin Pathol 2010; 63:879 - 83; http://dx.doi.org/10.1136/jcp.2009.073593; PMID: 20876318
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 2007; 131:861 - 72; http://dx.doi.org/10.1016/j.cell.2007.11.019; PMID: 18035408
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006; 126:663 - 76; http://dx.doi.org/10.1016/j.cell.2006.07.024; PMID: 16904174
- Kim JB, Greber B, Araúzo-Bravo MJ, Meyer J, Park KI, Zaehres H, et al. Direct reprogramming of human neural stem cells by OCT4. Nature 2009; 461:649 - 3; http://dx.doi.org/10.1038/nature08436; PMID: 19718018
- Wernig M, Meissner A, Foreman R, Brambrink T, Ku M, Hochedlinger K, et al. In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature 2007; 448:318 - 24; http://dx.doi.org/10.1038/nature05944; PMID: 17554336
- Kim K, Doi A, Wen B, Ng K, Zhao R, Cahan P, et al. Epigenetic memory in induced pluripotent stem cells. Nature 2010; 467:285 - 90; http://dx.doi.org/10.1038/nature09342; PMID: 20644535
- Lister R, Pelizzola M, Kida YS, Hawkins RD, Nery JR, Hon G, et al. Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stem cells. Nature 2011; 471:68 - 73; http://dx.doi.org/10.1038/nature09798; PMID: 21289626
- Shi Y, Desponts C, Do JT, Hahm HS, Schöler HR, Ding S. Induction of pluripotent stem cells from mouse embryonic fibroblasts by Oct4 and Klf4 with small-molecule compounds. Cell Stem Cell 2008; 3:568 - 74; http://dx.doi.org/10.1016/j.stem.2008.10.004; PMID: 18983970
- Feng B, Ng JH, Heng JC, Ng HH. Molecules that promote or enhance reprogramming of somatic cells to induced pluripotent stem cells. Cell Stem Cell 2009; 4:301 - 12; http://dx.doi.org/10.1016/j.stem.2009.03.005; PMID: 19341620
- Huangfu D, Osafune K, Maehr R, Guo W, Eijkelenboom A, Chen S, et al. Induction of pluripotent stem cells from primary human fibroblasts with only Oct4 and Sox2. Nat Biotechnol 2008; 26:1269 - 75; http://dx.doi.org/10.1038/nbt.1502; PMID: 18849973
- Doege CA, Inoue K, Yamashita T, Rhee DB, Travis S, Fujita R, et al. Early-stage epigenetic modification during somatic cell reprogramming by Parp1 and Tet2. Nature 2012; 488:652 - 5; http://dx.doi.org/10.1038/nature11333; PMID: 22902501
- Bhutani N, Brady JJ, Damian M, Sacco A, Corbel SY, Blau HM. Reprogramming towards pluripotency requires AID-dependent DNA demethylation. Nature 2010; 463:1042 - 7; http://dx.doi.org/10.1038/nature08752; PMID: 20027182
- Bultmann S, Morbitzer R, Schmidt CS, Thanisch K, Spada F, Elsaesser J, et al. Targeted transcriptional activation of silent oct4 pluripotency gene by combining designer TALEs and inhibition of epigenetic modifiers. Nucleic Acids Res 2012; 40:5368 - 77; http://dx.doi.org/10.1093/nar/gks199; PMID: 22387464
- Beltran AS, Blancafort P. Remodeling genomes with artificial transcription factors (ATFs). Methods Mol Biol 2010; 649:163 - 82; http://dx.doi.org/10.1007/978-1-60761-753-2_10; PMID: 20680834
- de Groote ML, Verschure PJ, Rots MG. Epigenetic Editing: targeted rewriting of epigenetic marks to modulate expression of selected target genes. Nucleic Acids Res 2012; 40:10596 - 613; http://dx.doi.org/10.1093/nar/gks863; PMID: 23002135
- Blancafort P, Segal DJ, Barbas CF 3rd. Designing transcription factor architectures for drug discovery. Mol Pharmacol 2004; 66:1361 - 71; http://dx.doi.org/10.1124/mol.104.002758; PMID: 15340042
- Beltran AS, Blancafort P. Reactivation of MASPIN in non-small cell lung carcinoma (NSCLC) cells by artificial transcription factors (ATFs). Epigenetics 2011; 6:224 - 35; http://dx.doi.org/10.4161/epi.6.2.13700; PMID: 20948306
- Beltran A, Parikh S, Liu Y, Cuevas BD, Johnson GL, Futscher BW, et al. Re-activation of a dormant tumor suppressor gene maspin by designed transcription factors. Oncogene 2007; 26:2791 - 8; http://dx.doi.org/10.1038/sj.onc.1210072; PMID: 17057734
- Beltran AS, Russo A, Lara H, Fan C, Lizardi PM, Blancafort P. Suppression of breast tumor growth and metastasis by an engineered transcription factor. PLoS One 2011; 6:e24595; http://dx.doi.org/10.1371/journal.pone.0024595; PMID: 21931769
- Lara H, Wang Y, Beltran AS, Juárez-Moreno K, Yuan X, Kato S, et al. Targeting serous epithelial ovarian cancer with designer zinc finger transcription factors. J Biol Chem 2012; 287:29873 - 86; http://dx.doi.org/10.1074/jbc.M112.360768; PMID: 22782891
- Beltran AS, Sun X, Lizardi PM, Blancafort P. Reprogramming epigenetic silencing: artificial transcription factors synergize with chromatin remodeling drugs to reactivate the tumor suppressor mammary serine protease inhibitor. Mol Cancer Ther 2008; 7:1080 - 90; http://dx.doi.org/10.1158/1535-7163.MCT-07-0526; PMID: 18483297
- Sera T. Zinc-finger-based artificial transcription factors and their applications. Adv Drug Deliv Rev 2009; 61:513 - 26; http://dx.doi.org/10.1016/j.addr.2009.03.012; PMID: 19394375
- Blancafort P, Beltran AS. Rational design, selection and specificity of artificial transcription factors (ATFs): the influence of chromatin in target gene regulation. Comb Chem High Throughput Screen 2008; 11:146 - 58; http://dx.doi.org/10.2174/138620708783744453; PMID: 18336208
- Stolzenburg S, Rots MG, Beltran AS, Rivenbark AG, Yuan X, Qian H, et al. Targeted silencing of the oncogenic transcription factor SOX2 in breast cancer. Nucleic Acids Res 2012; 40:6725 - 40; http://dx.doi.org/10.1093/nar/gks360; PMID: 22561374
- Rivenbark AG, Stolzenburg S, Beltran AS, Yuan X, Rots MG, Strahl BD, et al. Epigenetic reprogramming of cancer cells via targeted DNA methylation. Epigenetics 2012; 7:350 - 60; http://dx.doi.org/10.4161/epi.19507; PMID: 22419067
- Iyengar S, Farnham PJ. KAP1 protein: an enigmatic master regulator of the genome. J Biol Chem 2011; 286:26267 - 76; http://dx.doi.org/10.1074/jbc.R111.252569; PMID: 21652716
- Iyengar S, Ivanov AV, Jin VX, Rauscher FJ 3rd, Farnham PJ. Functional analysis of KAP1 genomic recruitment. Mol Cell Biol 2011; 31:1833 - 47; http://dx.doi.org/10.1128/MCB.01331-10; PMID: 21343339
- Frietze S, Lan X, Jin VX, Farnham PJ. Genomic targets of the KRAB and SCAN domain-containing zinc finger protein 263. J Biol Chem 2010; 285:1393 - 403; http://dx.doi.org/10.1074/jbc.M109.063032; PMID: 19887448
- Meilinger D, Fellinger K, Bultmann S, Rothbauer U, Bonapace IM, Klinkert WE, et al. Np95 interacts with de novo DNA methyltransferases, Dnmt3a and Dnmt3b, and mediates epigenetic silencing of the viral CMV promoter in embryonic stem cells. EMBO Rep 2009; 10:1259 - 64; http://dx.doi.org/10.1038/embor.2009.201; PMID: 19798101
- Epsztejn-Litman S, Feldman N, Abu-Remaileh M, Shufaro Y, Gerson A, Ueda J, et al. De novo DNA methylation promoted by G9a prevents reprogramming of embryonically silenced genes. Nat Struct Mol Biol 2008; 15:1176 - 83; http://dx.doi.org/10.1038/nsmb.1476; PMID: 18953337
- Hochedlinger K, Plath K. Epigenetic reprogramming and induced pluripotency. Development 2009; 136:509 - 23; http://dx.doi.org/10.1242/dev.020867; PMID: 19168672
- Kim JB, Zaehres H, Wu G, Gentile L, Ko K, Sebastiano V, et al. Pluripotent stem cells induced from adult neural stem cells by reprogramming with two factors. Nature 2008; 454:646 - 50; http://dx.doi.org/10.1038/nature07061; PMID: 18594515
- Kobolak J, Kiss K, Polgar Z, Mamo S, Rogel-Gaillard C, Tancos Z, et al. Promoter analysis of the rabbit POU5F1 gene and its expression in preimplantation stage embryos. BMC Mol Biol 2009; 10:88; http://dx.doi.org/10.1186/1471-2199-10-88; PMID: 19732419
- Beerli RR, Segal DJ, Dreier B, Barbas CF 3rd. Toward controlling gene expression at will: specific regulation of the erbB-2/HER-2 promoter by using polydactyl zinc finger proteins constructed from modular building blocks. Proc Natl Acad Sci U S A 1998; 95:14628 - 33; http://dx.doi.org/10.1073/pnas.95.25.14628; PMID: 9843940
- Urrutia R. KRAB-containing zinc-finger repressor proteins. Genome Biol 2003; 4:231; http://dx.doi.org/10.1186/gb-2003-4-10-231; PMID: 14519192
- Collins T, Stone JR, Williams AJ. All in the family: the BTB/POZ, KRAB, and SCAN domains. Mol Cell Biol 2001; 21:3609 - 15; http://dx.doi.org/10.1128/MCB.21.11.3609-3615.2001; PMID: 11340155
- Peng S, Maihle NJ, Huang Y. Pluripotency factors Lin28 and Oct4 identify a sub-population of stem cell-like cells in ovarian cancer. Oncogene 2010; 29:2153 - 9; http://dx.doi.org/10.1038/onc.2009.500; PMID: 20101213
- Topisirovic I, Kentsis A, Perez JM, Guzman ML, Jordan CT, Borden KL. Eukaryotic translation initiation factor 4E activity is modulated by HOXA9 at multiple levels. Mol Cell Biol 2005; 25:1100 - 12; http://dx.doi.org/10.1128/MCB.25.3.1100-1112.2005; PMID: 15657436
- Tachibana M, Matsumura Y, Fukuda M, Kimura H, Shinkai Y. G9a/GLP complexes independently mediate H3K9 and DNA methylation to silence transcription. EMBO J 2008; 27:2681 - 90; http://dx.doi.org/10.1038/emboj.2008.192; PMID: 18818694
- Hu T, Liu S, Breiter DR, Wang F, Tang Y, Sun S. Octamer 4 small interfering RNA results in cancer stem cell-like cell apoptosis. Cancer Res 2008; 68:6533 - 40; http://dx.doi.org/10.1158/0008-5472.CAN-07-6642; PMID: 18701476
- Gao MQ, Choi YP, Kang S, Youn JH, Cho NH. CD24+ cells from hierarchically organized ovarian cancer are enriched in cancer stem cells. Oncogene 2010; 29:2672 - 80; http://dx.doi.org/10.1038/onc.2010.35; PMID: 20190812
- Hu L, McArthur C, Jaffe RB. Ovarian cancer stem-like side-population cells are tumourigenic and chemoresistant. Br J Cancer 2010; 102:1276 - 83; http://dx.doi.org/10.1038/sj.bjc.6605626; PMID: 20354527
- Samardzija C, Quinn M, Findlay JK, Ahmed N. Attributes of Oct4 in stem cell biology: perspectives on cancer stem cells of the ovary. J Ovarian Res 2012; 5:37; http://dx.doi.org/10.1186/1757-2215-5-37; PMID: 23171809
- Mak VC, Siu MK, Wong OG, Chan KK, Ngan HY, Cheung AN. Dysregulated stemness-related genes in gynecological malignancies. Histol Histopathol 2012; 27:1121 - 30; PMID: 22806899
- Park JT, Chen X, Tropè CG, Davidson B, Shih IeM, Wang TL. Notch3 overexpression is related to the recurrence of ovarian cancer and confers resistance to carboplatin. Am J Pathol 2010; 177:1087 - 94; http://dx.doi.org/10.2353/ajpath.2010.100316; PMID: 20671266
- Gentilini O, Masullo M, Rotmensz N, Peccatori F, Mazzarol G, Smeets A, et al. Breast cancer diagnosed during pregnancy and lactation: biological features and treatment options. Eur J Surg Oncol 2005; 31:232 - 6; http://dx.doi.org/10.1016/j.ejso.2004.11.012; PMID: 15780556
- Keleher AJ, Theriault RL, Gwyn KM, Hunt KK, Stelling CB, Singletary SE, et al. Multidisciplinary management of breast cancer concurrent with pregnancy. J Am Coll Surg 2002; 194:54 - 64; http://dx.doi.org/10.1016/S1072-7515(01)01105-X; PMID: 11800340
- Schultz DC, Ayyanathan K, Negorev D, Maul GG, Rauscher FJ 3rd. SETDB1: a novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes Dev 2002; 16:919 - 32; http://dx.doi.org/10.1101/gad.973302; PMID: 11959841
- Yu HB, Kunarso G, Hong FH, Stanton LW. Zfp206, Oct4, and Sox2 are integrated components of a transcriptional regulatory network in embryonic stem cells. J Biol Chem 2009; 284:31327 - 35; http://dx.doi.org/10.1074/jbc.M109.016162; PMID: 19740739
- Zaret KS, Carroll JS. Pioneer transcription factors: establishing competence for gene expression. Genes Dev 2011; 25:2227 - 41; http://dx.doi.org/10.1101/gad.176826.111; PMID: 22056668
- Wang ZX, Teh CH, Chan CM, Chu C, Rossbach M, Kunarso G, et al. The transcription factor Zfp281 controls embryonic stem cell pluripotency by direct activation and repression of target genes. Stem Cells 2008; 26:2791 - 9; http://dx.doi.org/10.1634/stemcells.2008-0443; PMID: 18757296
- Mandell JG, Barbas CF 3rd. Zinc Finger Tools: custom DNA-binding domains for transcription factors and nucleases. Nucleic Acids Res 2006; 34:Web Server issue W516-23; http://dx.doi.org/10.1093/nar/gkl209; PMID: 16845061