Abstract
DACT2 (Dapper, Dishevelled-associated antagonist of β-catenin homolog 2) is a member of the DACT family involved in the regulation of embryonic development. Human DACT2 is localized on 6q27, a region of frequent loss of heterozygosity in human cancers. However, the regulation of DACT2 expression and function in hepatocellular carcinoma (HCC) remains unclear. In this study, genetic and epigenetic changes of DACT2 were analyzed in HCC cell lines and primary cancer. We found no single-nucleotide polymorphism (SNP) associated with HCC. Promoter region methylation was correlated with loss or reduction of DACT2 expression, and restoration of DACT2 expression was induced by 5-aza-2’-deoxycytidine (5-AZA) in HCC cell lines. Promoter region methylation was found in 54.84% of primary HCC. Reduction of DACT2 expression was associated with promoter hypermethylation, and expression of DACT2 was inversely related to β-catenin expression in primary HCC. DACT2 suppressed cell proliferation, induced G2-M arrest in cell lines and inhibited tumor growth in xenograft nude mice. The transcriptional activity of TCF-4 and the expression of Wnt signaling downstream genes were suppressed by DACT2 re-expression and reactivated by depletion of DACT2. In conclusion, DACT2 is frequently methylated in HCC and its expression is regulated by promoter hypermethylation. DACT2 suppresses HCC by inhibiting Wnt signaling in human HCC.
Introduction
In men, hepatocellular carcinoma (HCC) is the fifth most frequently diagnosed cancer worldwide and the second cause of global cancer death. In women, it is the seventh most commonly diagnosed cancer and the sixth leading cause of cancer death.Citation1 Carcinogenesis is a multi-step process with cumulative genetic and epigenetic changes involving oncogenes and tumor suppressor genes.Citation2-Citation6 An increasing number of publications are focusing on identifying novel genes regulated by DNA methylation, histone modification and miRNAs.Citation7-Citation12 These studies are mainly focused on elucidating the mechanisms of inactivation of tumor suppressors in different signaling pathways in order to find new therapeutic strategies in human cancer, including HCC. Wnt signaling plays an important role in the physiology, development, cell differentiation, proliferation and growth.Citation13 Abnormal activation of Wnt signaling is a major driving force in HCC.Citation14-Citation16
Dapper, a Dishevelled-associated antagonist of β-catenin (DACT), was isolated by a screen for proteins interacting with Dishevelled, a key factor in the Wnt signaling. Dapper and Dishevelled were co-localized intracellularly and formed a complex with Axin, GSK3 and β-catenin.Citation17 Human DACT1 and DACT2 were identified by Katoh et al. in 2003.Citation18 Human and murine DACT3 were both identified by Fisher et al.Citation19 DACT1 has been reported frequently to be methylated in HCC, and DACT3 has been found to be regulated by histone modifications in colorectal cancer.Citation12,Citation20 Human DACT2 is localized in chromosome 6q27, a region of frequent loss of heterozygosity in human cancers.Citation18,Citation21-Citation27 However, the regulation of DACT2 expression and its function in human HCC remains unknown.
In this study, we first analyzed genetic and epigenetic changes of DACT2, and then studied its expression and function in hepatic carcinogenesis in vitro and in vivo.
Results
No single-nucleotide polymorphism (SNP) in the DACT2 gene is associated with HCC
The sequencing of the full length cDNA and genomic DNA of DACT2 in seven hepatic cancer cell lines and one immortalized hepatocyte cell line (LO2) revealed five SNP in exon 4, an important functional region also known as PDZ (post synaptic density-95/discs large/zonula occludens-1) binding domain.Citation28 Although no new mutations were discovered, four of the above SNPs were found both in patients with HCCs and in healthy controls. The respective locations and frequencies of these SNPs in both patients with HCCs and in healthy controls are as follows: 26.25% vs. 23.10% for A/G (rs6925614), 2.50% vs. 1.28% for T/C (rs79931308), 15.00% vs. 15.38% for A/C (rs10945501) and 1.25% vs. 1.28% for G/T (rs73789362). No significant differences were found in SNPs between HCCs patients and healthy individuals (p > 0.05).
DACT2 is silenced by promoter hypermethylation in HCC cell lines
DACT2 was silenced in the HepG2 cell line and reduced in cell lines SNU182, BEL7402, SMMC7721 and SNU449. DACT2 was normally expressed in PLC/PRF/5, 97H and in the immortalized cell line (LO2) (). To investigate if silencing of DACT2 is associated with promoter region hypermethylation, we first analyzed the CpG island of DACT2 DNA sequence using a CpG Island search program (http://cpgislands.usc.edu). One CpG island was found in the promoter region (). Then DACT2 promoter region methylation was analyzed by MSP and bisulfite sequencing (BSSQ). Complete methylation was found in the HepG2 cells, and partial methylation was observed in the SNU182, BEL7402, SMMC7721 and SNU449 cell lines. No methylation was detected in LO2, PLC/PRF/5 and 97H cell lines (). The methylation density within DACT2 promoter region was characterized and validated by BSSQ (). Bisulfite sequencing of 10 individual clones of PCR products from HepG2 revealed dense methylation of CpGs within the promoter region. The mixed methylation pattern of CpGs observed with BSSQ in the SNU182 cell line may represent both methylated and unmethylated alleles or both methylated and unmethylated clonal subpopulations within cultured cells. No methylation was found by BSSQ in PLC/PRF/5 and LO2. These results indicate that our MSP assays results accurately represent DACT2 promoter region methylation status in these cell lines.
Figure 1.DACT2 is silenced by promoter region hypermethylation in HCC cell lines.(A) Expression of DACT2 was analyzed by semiquantitative RT-PCR in HCC cell lines and one immortalized hepatocyte cell line (LO2). (-) 5-AZA untreated; (+) 5-AZA treated; GAPDH was used as an internal control for RT-PCR. (B) Distribution of CpG sites in the promoter region of DACT2 and the location of the MSP primers as well as BSSQ region are shown. (C) DACT2 methylation was determined by MSP in HCC cell lines and LO2. IVD, in vitro methylated DNA (methylation control); NL, normal blood lymphocyte DNA (unmethylated control); M, methylated band; U, unmethylated band. (D) Promoter region methylation status of DACT2 was analyzed by BSSQ in three HCC cell lines. Open circles denote unmethylated CpG site and filled circles represented methylated CpG site. The region amplified by MSP is indicated by arrows. TSS, transcription start site.
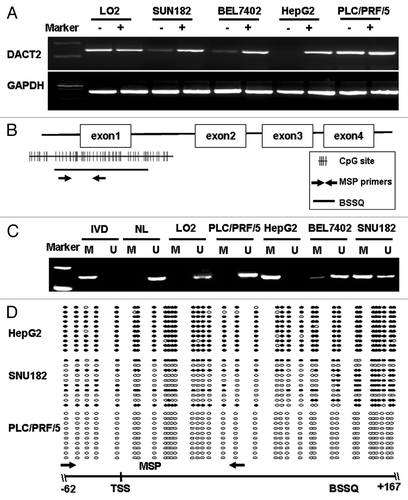
Concomitant loss of DACT2 expression together with promoter region complete methylation was found in HepG2 cells. Normal expression without concomitant methylation was observed in LO2, PLC/PRF/5 and 97H cells. Partial methylation and reduced expression were detected in SNU182, BEL7402, SMMC7721 and SNU449 cell lines. These results indicate that promoter region methylation is correlated with DACT2 silencing. DACT2 expression was restored after 5-AZA treatment in HepG2 cells, and increased expression was observed in the SNU182 and BEL7402 cell lines. All of the above results demonstrated that DACT2 expression was regulated by promoter region hypermethylation.
DACT2 is frequently methylated in primary HCCs
DACT2 promoter region hypermethylation was not limited to cultured HCC cell lines. Frequent methylation was found in primary HCC (). In 62 HCCs, 34 cases (54.84%) were methylated and 28 cases (45.16%) were unmethylated. No association was found between DACT2 methylation and clinicopathological variables such as age, gender, hepatitis B/C virus infection, cirrhosis, AFP levels, tumor size or tumor stage in HCCs ().
Figure 2.DACT2 expression is associated with promoter hypermethylation in primary HCCs. (A) Representative MSP results of DACT2 in primary HCCs. (B) DACT2 expression in a representative HCC case. Left image: adjacent non-tumor tissue (100×) and the indicated fields enlarged (400×). Right image: tumor tissue (100×) and the indicated fields enlarged (400×). (C) DACT2 expression scores are shown as box plots, the horizontal lines represent the median score; the bottom and top of the boxes represent the 25th and 75th percentiles, respectively; the vertical bars represent the range of data. Expression of DACT2 was different between non-tumor tissues and tumor tissues in 62 matched primary HCCs. **p < 0.01. (D) The correlation of DACT2 hypermethylation and expression level were analyzed in 62 matched primary HCCs. *p < 0.05. (E) β-catenin expression in the representative methylated case (400×). β-catenin expression is positive mostly in the membranes of hepatic cells (left image) and in the cytoplasm and nucleus in tumor cells (right image).
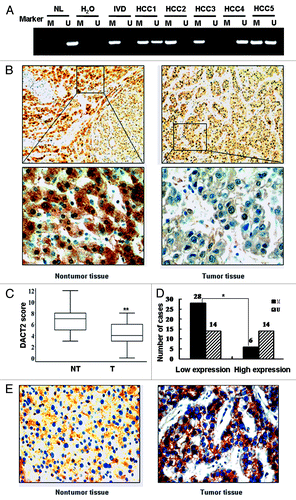
Table 1. Association of clinical factors with DACT2 methylation and expression in HCC patients
DACT2 expression is associated with promoter hypermethylation in primary HCCs
DACT2 expression was analyzed by IHC in tumor tissues and adjacent non-tumor tissues. DACT2 was expressed in the cytoplasm of hepatic cells and its expression was reduced significantly in tumor tissues compared with adjacent tissues (p < 0.01) (). Reduced DACT2 expression was related to younger age (p < 0.01) and high serum levels of AFP (p < 0.05) patients (). These results suggest that DACT2 may be a potential prognosis marker.
The association of DACT2 expression and methylation status was analyzed in 62 HCC patients. Reduced expression of DACT2 was found in 42 cases of tumor tissue, of which 28 cases were methylated (66.67%). Twenty cases were normally expressed, of which 6 cases were methylated (30%). Reduced DACT2 expression was significantly related to promoter methylation (p < 0.05, ). This suggests that DACT2 expression may be regulated by promoter region methylation in primary HCC.
To evaluate whether activation of Wnt signaling was related to DACT2 expression, β-catenin expression was analyzed by IHC in DACT2 reduction cases. In cancer tissues, positive cytoplasm and nucleus β-catenin staining was found in 25 cases (59.52%). In adjacent tissues, β-catenin expression was mainly located on the membrane (31/42, 73%) (). The expression of DACT2 was associated with β-catenin location in HCC (p < 0.05).
HCC cell growth is suppressed by DACT2 re-expression
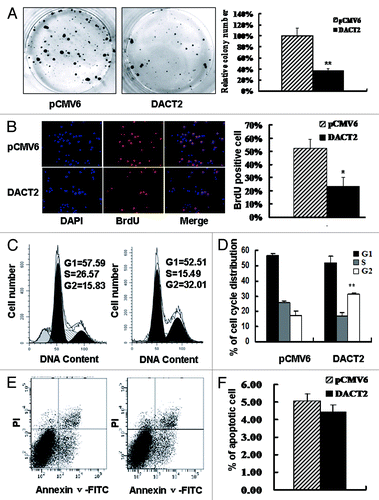
Frequent loss of DACT2 expression in HCCs suggests that it may be a potential tumor suppressor. To evaluate the effect of DACT2 on cancer cell proliferation, colony formation assay was employed. DACT2 expression resulted in significant suppression of long-term cell growth in colony formation assays, as much as 60% (). Concomitantly, the inhibitory effect of DACT2 was evaluated by BrdU incorporation assay. HepG2 growth was inhibited about 30% by DACT2 ().
Figure 3. The effect of DACT2 on cell proliferation, cell cycle and apoptosis in vitro. (A) The effect of DACT2 on HCC cell proliferation was evaluated by colony formation assay. Left panel: Colony formation results of transfected with pCMV6 (empty vector) or DACT2 expression vector in HepG2 cells. Right panel: Quantitative analysis of colony numbers in different group. **p < 0.01. (B) BrdU incorporation was analyzed in pCMV6 or DACT2 expression vector transfected HepG2 cells. Left panel: cell nuclei were counterstained by BrdU and DAPI. Right panel: quantitative analysis of BrdU positive cells in different group. *p < 0.05. (C) Representative results of cell cycle distribution for pCMV6 (left panel) or DACT2 expression vector (right panel) transfected in HepG2 cells. Cycle distributions were measured by propidium iodide (PI) staining followed by flow cytometry after transfection for 48 h. (D) Quantitative analysis of cell cycle distribution in DACT2 expression and control group. **p < 0.01. (E) No effect was found on apoptosis after expression of DACT2 in HepG2 cell line. HepG2 cells were transfected with pCMV6 (left panel) or DACT2 expression vector (Right panel). 48 h serum starvation was performed 24 h after transfection. Then Annexin V-FITC/PI double staining was performed. Annexin V-positive and PI-negative staining cells are indicated as apoptotic cells. (F) Quantitative analysis of cell apoptosis.
G2-M arrest is induced by re-expression of DACT2
The effect of DACT2 on the cell cycle was evaluated by flow cytometry. The ratio of G2 phase was increased after re-expression of DACT2 in HepG2 cells (G1: 56.60 ± 1.72 vs 51.70 ± 4.27; S: 25.95 ± 1.07 vs 16.64 ± 2.50; G2:17.45 ± 2.81 vs 31.0 ± 1.27) (p < 0.01) (). These results indicate that DACT2 induces G2-M phase arrest in HCC.
DACT2 has no effects on cell apoptosis
To investigate the effects of DACT2 on apoptosis, an annexin V-FITC assay was employed in HepG2 cells. No difference was found between the DACT2 expression group and control group (11.98% ± 2.19 vs. 13.60% ± 1.67, p > 0.05) (). These data suggest that DACT2 is not involved in apoptosis in HCC cells.
Tumor growth is retarded by DACT2 in vivo
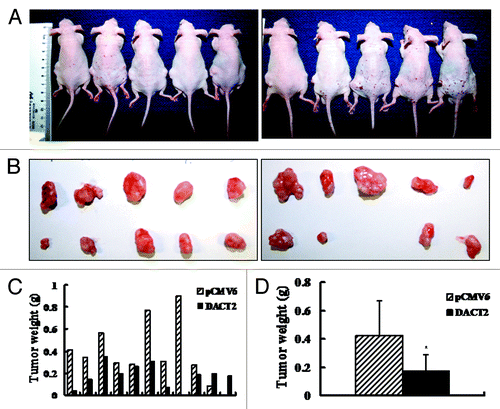
Mice burdened with subcutaneous tumors were shown in . shows that the volume is significantly different in DACT2 expressed and unexpressed HepG2 groups (251.09 ± 174.24 mm3 vs. 533.20 ± 370.86 mm3, p < 0.05). As shown in , the tumor weight is reduced in DACT2-expressing nude mice compared with an empty vector group (0.18 ± 0.11 g vs. 0.42 ± 0.25 g; p < 0.05). The results indicate that DACT2 inhibits tumor growth in vivo.
Figure 4. Tumor growth was inhibited by DACT2 in vivo. (A) Nude mice inoculated with HepG2 cells expressing or not expressing DACT2. Left rear flank: empty vector group. Right rear flank: DACT2-expressing group (n = 10/group). (B) Tumors in the upper panel are from the empty vector group and tumors in the lower panel are from the DACT2-expressing group. (C) Histogram represents tumor weights of each mouse in the two groups. (D) Tumor weights are significantly different between DACT2-expressing and empty vector groups. *p < 0.05.
DACT2 is an inhibitor of Wnt signaling
DACT2 was reported to be involved in Wnt signaling during zebra fish and mice development. To explore the effects of DACT2 on Wnt signaling, promoter-luciferase activity assays were employed in this study. The transcriptional activity of TCF-4 was inhibited in both wild type and mutant β-catenin vectors groups by the re-expression of DACT2 (). Western blotting shows that the expression of TCF-4/β-catenin downstream targets, c-Myc and cyclin D1, was decreased after re-expression of DACT2 in HepG2 cells (). These results strongly suggest that DACT2 is a Wnt signaling inhibitor in HCC.
Figure 5. DACT2 is a Wnt signaling inhibitor. (A) The transcriptional activity of TCF-4 was inhibited by DACT2. Left panel: DACT2 was co-transfected with wt β-catenin and OT into HepG2 cells. Right panel: DACT2 was co-transfected with mut β-catenin and OT into HepG2 cells. OF acted as a negative control reporter and its transcriptional activity was defined as 1. *p < 0.05. (B) Expression of c-Myc and cyclin D1 was analyzed by western blot in DACT2-expressing or -not expressing HepG2 cells. Actin was used as an internal control. (C) The transcriptional activity of TCF-4 was increased after two individual siRNAs targeting DACT2 vectors were transfected into PLC/PRF/5 cells. *p < 0.05. (D) Knockdown of DACT2 increases the expression of c-Myc and cyclin D1 in PLC/PRF/5 cells. Actin was used as internal reference.
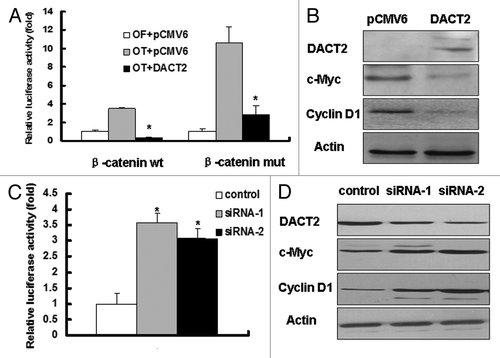
To further analyze the effect of DACT2 on Wnt signaling, DACT2 was knocked down by RNA interference (RNAi) in PLC/PRF/5 cells. As shown in , the transcriptional activity of TCF-4 was increased in DACT2-depleted cells, compared with control group, and the expression of Wnt signaling downstream genes was increased when depletion of DACT2 in PLC/PRF/5 cells. These results suggest that DACT2 is a Wnt signaling inhibitor in HCC.
Discussion
Abnormal activation of Wnt signaling is a major driving force in cancer, which can be initiated by genetic or epigenetic changes.Citation4,Citation29 Frequent methylation of Wnt signaling antagonists suggest an important role for the activation of this pathway during carcinogenesis.Citation5,Citation30
DACT2 is located on human chromosome 6q27, a region frequently associated with loss of heterozygosity in human cancers.Citation18,Citation21-Citation27
DACT genes encode a small family of vertebrate intracellular proteins that can regulate intercellular signaling pathways by a conserved leucine zipper motif near the N-terminus and a binding motif for PDZ domain at the C-terminus.Citation17,Citation18,Citation31DACT1 and DACT2 were discovered by two independent groups who were screening partners of the Dvl scaffold protein, which is central to the developmentally and clinically important Wnt signaling.Citation31-Citation36 The initial functional analyses of DACT2 were based on its overexpression and morpholino-based knockdown technologies in Xenopus laevis and zebrafish.Citation37,Citation38
In this study, we describe for the first time that DACT2 is frequently absent or downregulated in HCC cell lines, and is also significantly reduced in primary HCC samples. We first sequenced DACT2 in primary HCC and cell lines and found no mutation related to HCC. This suggests that genetic changes may not play important roles in HCC carcinogenesis. It was previously reported that DACT2 is frequently inactivated by DNA methylation in colorectal cancer cell lines (RKO and HT29).Citation12 Our previous study in lung cancer also reported that the DACT2 gene was silenced by promoter region methylation.Citation39 To determine whether hypermethylation was responsible for the silencing of DACT2 in HCC, promoter region methylation was analyzed in HCC cell lines and primary cancer. Our data suggest that DACT2 is regulated by promoter region hypermethylation in HCC. Further analysis indicates that reduction of DACT2 expression was related to younger age and high level of AFP in serum. These results suggest that methylation of DACT2 may serve as a diagnostic tool and prognostic marker in HCC.
It has been reported that DACT2 binds to the TGF-β receptors ALK5 and ALK4, accelerating lysosomal degradation of these receptors in zebrafish.Citation38 Although recent studies showed that murine DACT proteins formed a weak complex with Alk5 in HEK293T cells, no such complex can be detected between Alk4 or Alk5 and DACT2 protein.Citation36 DACT2 strongly co-immunoprecipitated with β-catenin or δ-catenin in HEK293T cells, as well as formed even stronger complexes with CDK1δ/ε, Dvl or Vangl family members.Citation36 This report suggests that DACT2 is a Wnt signaling inhibitor. Our study found that HCC proliferation was suppressed by DACT2 both in vitro and in vivo. In addition, Wnt signaling activity was inhibited by DACT2 in HCC cells. These results suggest that DACT2 inhibits human HCC by inactivating Wnt signaling.
In conclusion, DACT2 is silenced by promoter region hypermethylation in human hepatic cancer and its methylation may serve as a detection marker for HCC. DACT2 suppressed cell proliferation, induced G2-M arrest in cell cycle and inhibited tumor growth in nude mice. DACT2 also inhibited TCF-4 transcriptional activity and its downstream targets (c-Myc and cyclin D1) in HCC. Based on these findings and its important human chromosomal localization, DACT2 may be a tumor suppressor in HCC.
Materials and Methods
Ethics statement
For the use of clinical materials for research purposes, prior patients’ consent and approval were obtained from Institutional Review Board of the Chinese PLA General Hospital.
Cell lines and tissue samples
Eight HCC cell lines (LO2, SNU182, BEL7402, HepG2, PLC/PRF/5, SMMC7721, SNU449 and 97H) were used in this study. Cell lines were cultured at 37°C in an atmosphere containing 5% CO2 in 90% RPMI 1640 (Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum. Cells were passaged 1:3 once 80% confluence (approximately 1 × 106 cells) was reached on a 75 cm2 culture flask (NEST Biotechnology).
Sixty-two paired tumor tissue samples of primary HCC and their adjacent non-tumor tissues were obtained from surgical resected HCC patients at the Chinese PLA General Hospital, Beijing. For this study, all of the tumor and non-tumor tissue were re-examined and confirmed by pathologists.
RNA isolation and semi-quantitative RT-PCR
RNA Isolation and semi-quantitative RT-PCR. Total RNA was isolated using the RNeasy mini kit (Qiagen). First strand cDNA (cDNA) was synthesized using the Superscript II-reverse transcriptase kit (Invitrogen). Expression of DACT2 mRNA (mRNA) was determined by RT-PCR using the LightCycler system (Roche Diagnostics,). PCR amplification of DACT2 was performed using primers: 5′-GGCTGAGACAACAGGACATCG-3′ (forward) and 5′-GACCGTCGCTCATCTCGTAAAA-3′ (reverse). The primer set for DACT2 was designed to span intronic sequences between exons in order to control genomic DNA contamination. A total of 35 cycles of amplification were performed for each of the RT-PCR experiments. As an internal control, GAPDH was amplified with 25 cycles to ensure cDNA quality and quantity for each RT-PCR. Amplified products were analyzed on 1.5% agarose gel.
Methylation-specific PCR (MSP) and bisulfite sequencing
Genomic DNA from cell lines and tissue samples was prepared using the proteinase-K method. After chloroform/phenol extraction, DNA was precipitated in ethanol and dissolved in low TE buffer and stored at -20°C. Genomic DNA from HCC tissues and cell lines was bisulfite modified as previously described.Citation40 MSP primers were designed according to genomic sequences flanking the presumed transcription start site (TSS). Primer sequences were oligo-synthesized (Invitrogen) to allow MSP to detect bisulfite-induced changes affecting unmethylated (U) and methylated (M) alleles. MSP of DACT2 was performed using primers: 5′-GATTTTAGTTTATTTTGGCGATTTGC-3′ (M-forward); 5′-CACATCTCCCGAACAAAATCCCG-3′ (M-reverse); 5′-TAGATTTTAGTTTATTTTGGTGATTTGT-3′ (U-forward) and 5′- TCCACATCTCCCAAACAAAATCCCA -3′ (U-reverse). Each MSP reaction incorporated approximately 100 ng of bisulfite-treated DNA, 25 pmoles of each primer, 100 pmoles dNTPs, 2.5 μl 10 × PCR buffer, and 1 unit of Taq Polymerase (Invitrogen) in a final reaction volume of 25 μl. Cycle conditions were: 95°C × 5 min, 1 cycle; 35 cycles × (95°C × 30 sec, 60°C × 30 sec, 72°C × 30 sec); 72°C × 5 min, 1 cycle. Each PCR assay included a methylated control (in vitro methylated DNA, IVD), an unmethylated control (normal blood lymphocyte DNA, NL) and a negative control (water). MSP products were analyzed using 2% agarose gel electrophoresis.
Bisulfite-treated DNA was subjected to PCR using primers flanking the targeted MSP regions above. Sequencing primers were as follows: 5′-TGGTTATAGATTTTAGTTTATTTTGG-3′ (forward) and antisense 5′-CAACCCCTACAACTCCTACAAC-3′ (reverse). PCR cycle conditions were as follows: 95°C × 5 min, 1 cycle; 35 cycles × (95°C × 30sec, 58°C × 30sec, 72°C × 40 sec); 72°C × 5 min, 1 cycle. PCR products were gel purified and cloned into pCR2.1 vector according to the manufacturer’s instructions (Invitrogen). Sequencing was performed as previously reported.Citation5, Citation41
5-aza-2’-deoxycytidine (5-AZA) treatment
Cell lines (HepG2, SNU182, BEL7402, PLC/PRF/5 and LO2) were split to a low density (30% confluence) 12 h before treatment. Cells were treated with 5-AZA (Sigma) at a concentration of 2 μM. Growth medium, conditioned with 5-AZA at 2 μM, was exchanged every 24 h for 96 h. At the end of the treatment, RNA was extracted from the cells as described above.
Immunohistochemistry staining
Rabbit anti-DACT2 antibody (Prosci) and mouse anti-β-catenin antibody (ZSGB Biotech.) were employed. Immunohistochemistry (IHC) was performed on 4μm thick serial sections derived from formaldehyde fixed paraffin wax embedded tumor tissue blocks. After de-paraffinization and rehydration, endogenous peroxidase activity was blocked for 30 min in methanol containing 0.3% hydrogen peroxide. The slides were then incubated with anti-DACT2 antibodies (1:1600 dilution) or with mouse anti-β-catenin antibody (1:400) overnight at 4°C in a humidified chamber. IHC was performed on the tissue sections according to the Polink-2 plus® Polymer HRP Detection System (ZSGB Biotech). Hematoxylin was used for counterstaining. The expression of DACT2 and β-catenin was evaluated according to previous report.Citation42
Construction of DACT2 expression vector and isolation of DACT2 expressed cells
The expression construct for DACT2 was generated by cloning a PCR-amplified full-length human DACT2 cDNA fragment (GenBank accession number NM_214462) into pCMV6-AC-GFP or pCMV6-Entry-myc (Origene Technologies, Inc.). The vector was verified by DNA sequencing and western Blotting. Transient transfection was performed using FuGENE 6 (Roche Applied Science) according to the manufacturer’s instructions. GFP positive cells were isolated by flow cytometry (Becton Dickinson) 48 h after transfection.
Western blotting
Cell lysates were collected and western blotting was performed. The antibodies used included rabbit anti-DACT2 antibodies (Prosci) and monoclonal antibodies against cyclin D1 (Santa Cruz Biotechnology) as well as c-Myc and actin (Bioworld Technology)
Colony formation assay
HepG2 cells were transfected with either an empty or a DACT2 expression vector using FuGENE HD (Roche Applied Science) according to manufacturer’s instructions. 60% GFP positive cells were isolated by flow cytometry 48 h after transfection. Cells were collected and reseeded in triplicate at 1,500 cells per well in 6-well plates. Growth medium, conditioned with G418 (Invitrogen) at 500 μg/mL, was exchanged every 24 h. After 10–14 d, cells were fixed with 75% ethanol for 30 min, stained with 0.2% crystal violet for visualization, and counted.
BrdU incorporation analysis
HepG2 cells, transfected with either an empty or a DACT2 expression vector, were seeded in 24-well plates. Ten milimolar BrdU was added in growth medium and incubated at 37°C for 2 h. Cells were stained with mouse anti-BrdU antibody and DAPI for double labeling. The BrdU (red) /DAPI (blue) positively staining cells are indicated as positive BrdU incorporation.
Cell cycle analysis
DACT or empty vector was transfected into HepG2 cells using FuGENE. Forty-eight hours after transfection, cells were harvested, washed with phosphate-buffered saline (PBS) and fixed with ice-cold 70% ethanol at -20°C overnight. Samples were then washed with PBS and stained with propidium iodide (Sigma) containing RNase A (Sigma) for 30 min at 37°C. Cell cycle distribution in different phases was determined using flow cytometry.
In vivo tumor genesis assay
Six-week-old female nu/nu mice were bred under specified pathogen-free conditions. HepG2 cells were transiently transfected with either an empty or a DACT2 expression vector using FuGENE HD and GFP positive cells were isolated by flow cytometry 48 h after transfection. Cells (3 × 106) were diluted in 100 μl PBS and injected subcutaneously into the left or right rear flank of the mice. When tumors reached approximately 1.0 cm in size, the mice were sacrificed and the volume as well as the weight of the tumors were measured. Tumor volume (mm3) was estimated by measuring the longest and shortest diameter of the tumor and calculated with the formula: tumor volume = (length) × (width)2/2. All experimental procedures were approved by the Animal Ethics Committee of the Chinese PLA General Hospital, Beijing.
Dual-luciferase reporter assay
To explore the effect of DACT2 on Wnt signaling, DACT2 was co-transfected with TCF-4 reporter, pTOPFlash (OT), and β–catenin wide type (wt) or β-catenin mutant (mut) vector into HepG2 cells, pFOPFlash (OF) acted as a negative control reporter.Citation43 Cell lysates were collected and luciferase enzymatic luminescent activity was measured 48 h after transfection according to the manufacturer’s instructions (GLOMAX luminometer, Dual Luciferase Reporter Assay system, Promega).
RNA interference
Two selected siRNAs targeting DACT2 and RNAi Negative Control Duplex were used in this study. The sequences are as follows: siRNA-1 (sense sequence: 5′-CCAGCUGUCCUGAGUCUAATT-3′ and antisense sequence: 5′-UUAGACUCAGGACAGCUGGTT-3′); siRNA-2 (sense sequence: 5′-GUCGGUUGAUGAGACUACUTT-3′ and antisense sequence: 5′-AGUAGUCUCAUCAACCGACTT-3′). RNAi Negative Control Duplex (sense sequence: 5′-UUCUCCGAACGUGUCACGUTT-3′; and antisense sequence: 5′-ACGUGACACGUUCGGAGAATT-3). Fifty to sixty percent confluent PLC/PRF/5 cells were transfected with 50 nM of siRNAs using Lipofectamine 2000 (Invitrogen) following the manufacturer’s direction.
Statistical analysis
We evaluated the relationship between methylation status in human hepatocellular carcinomas and clinicopathologic characteristics using the Pearson’s chi-square test or the Fisher’s exact test for independence for dichotomous variables as appropriate. Continuous variables were analyzed with Student’s t-test. Results were judged to be statistically significant at p < 0.05 and all p values were two-sided. All analyses were done using SPSS PASW Statistics 18.0.
| Abbreviations: | ||
| HCC | = | hepatocellular carcinoma |
| DACT2 | = | Dapper, a Dishevelled-associated antagonist of β-catenin, homolog 2 |
| MSP | = | methylation specific polymerase chain reaction |
| BSSQ | = | Bisulfite sequencing |
| GAPDH | = | glyceraldehyde-3-phosphate dehydrogenase |
| TSS | = | transcription start site |
| M | = | methylated |
| U | = | unmethylated |
| 5-AZA | = | 5-aza-2’-deoxycytidine |
| NL | = | normal blood lymphocyte DNA |
| IVD | = | in vitro methylated DNA |
| PDZ | = | Post synaptic density-95/Discs large/Zonula occludens-1 |
| PBS | = | phosphate-buffered saline |
| AJCC | = | American Joint Committee on Cancer |
| AFP | = | alpha-fetoprotein |
| ALTSG | = | American Liver Tumor Study Group |
| BrdU | = | bromodeoxyuridine |
| TCF | = | T-cell factor |
| LEF | = | lymphoid enhancer factor |
| NT | = | non-tumor tissue |
| T | = | tumor tissue |
| SNP | = | single-nucleotide polymorphism |
Acknowledgments
This work was supported by grants from the National Basic Research Program of China (973 Program No. 2012CB934002, 2010CB912802); National High-tech R&D Program of China (863 Program No. SS2012AA020314, SS2012AA020821, SS2012AA020303); National Key Scientific instrument Special Programme of China (Grant No. 2011YQ03013405); National Science Foundation of China (Grant No. 81121004, 81071953 and 81161120432).
Disclosure of Potential Conflicts of Interest
J.G.H. is a consultant to MDxHealth. The other authors declare no conflict of interest.
References
- Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin 2011; 61:69 - 90; http://dx.doi.org/10.3322/caac.20107; PMID: 21296855
- Guo M, Ren J, House MG, Qi Y, Brock MV, Herman JG. Accumulation of promoter methylation suggests epigenetic progression in squamous cell carcinoma of the esophagus. Clin Cancer Res 2006; 12:4515 - 22; http://dx.doi.org/10.1158/1078-0432.CCR-05-2858; PMID: 16899597
- Zhang XM, Guo MZ. The value of epigenetic markers in esophageal cancer. Front Med China 2010; 4:378 - 84; http://dx.doi.org/10.1007/s11684-010-0230-3; PMID: 21107750
- Baylin SB, Ohm JE. Epigenetic gene silencing in cancer - a mechanism for early oncogenic pathway addiction?. Nat Rev Cancer 2006; 6:107 - 16; http://dx.doi.org/10.1038/nrc1799; PMID: 16491070
- Jia Y, Yang Y, Liu S, Herman JG, Lu F, Guo M. SOX17 antagonizes WNT/β-catenin signaling pathway in hepatocellular carcinoma. Epigenetics 2010; 5:743 - 9; http://dx.doi.org/10.4161/epi.5.8.13104; PMID: 20716954
- Guo M, Liu S, Lu F. Gefitinib-sensitizing mutations in esophageal carcinoma. N Engl J Med 2006; 354:2193 - 4; http://dx.doi.org/10.1056/NEJMc052698; PMID: 16707764
- Guo M, House MG, Suzuki H, Ye Y, Brock MV, Lu F, et al. Epigenetic silencing of CDX2 is a feature of squamous esophageal cancer. Int J Cancer 2007; 121:1219 - 26; http://dx.doi.org/10.1002/ijc.22828; PMID: 17534889
- Guo M, House MG, Akiyama Y, Qi Y, Capagna D, Harmon J, et al. Hypermethylation of the GATA gene family in esophageal cancer. Int J Cancer 2006; 119:2078 - 83; http://dx.doi.org/10.1002/ijc.22092; PMID: 16823849
- Liu W, Li X, Chu ES, Go MY, Xu L, Zhao G, et al. Paired box gene 5 is a novel tumor suppressor in hepatocellular carcinoma through interaction with p53 signaling pathway. Hepatology 2011; 53:843 - 53; http://dx.doi.org/10.1002/hep.24124; PMID: 21319196
- Wong CM, Wong CC, Lee JM, Fan DN, Au SL, Ng IO. Sequential alterations of miRNA expression in hepatocellular carcinoma development and venous metastasis. Hepatology 2011; 55:1453 - 61; http://dx.doi.org/10.1002/hep.25512
- Wang Y, Lee CG. Role of miR-224 in hepatocellular carcinoma: a tool for possible therapeutic intervention?. Epigenomics 2011; 3:235 - 43; http://dx.doi.org/10.2217/epi.11.5; PMID: 22122284
- Jiang X, Tan J, Li J, Kivimäe S, Yang X, Zhuang L, et al. DACT3 is an epigenetic regulator of Wnt/beta-catenin signaling in colorectal cancer and is a therapeutic target of histone modifications. Cancer Cell 2008; 13:529 - 41; http://dx.doi.org/10.1016/j.ccr.2008.04.019; PMID: 18538736
- Klaus A, Birchmeier W. Wnt signalling and its impact on development and cancer. Nat Rev Cancer 2008; 8:387 - 98; http://dx.doi.org/10.1038/nrc2389; PMID: 18432252
- Kim M, Lee HC, Tsedensodnom O, Hartley R, Lim YS, Yu E, et al. Functional interaction between Wnt3 and Frizzled-7 leads to activation of the Wnt/beta-catenin signaling pathway in hepatocellular carcinoma cells. J Hepatol 2008; 48:780 - 91; http://dx.doi.org/10.1016/j.jhep.2007.12.020; PMID: 18313787
- Bengochea A, de Souza MM, Lefrançois L, Le Roux E, Galy O, Chemin I, et al. Common dysregulation of Wnt/Frizzled receptor elements in human hepatocellular carcinoma. Br J Cancer 2008; 99:143 - 50; http://dx.doi.org/10.1038/sj.bjc.6604422; PMID: 18577996
- Lee HC, Kim M, Wands JR. Wnt/Frizzled signaling in hepatocellular carcinoma. Front Biosci 2006; 11:1901 - 15; http://dx.doi.org/10.2741/1933; PMID: 16368566
- Cheyette BN, Waxman JS, Miller JR, Takemaru K, Sheldahl LC, Khlebtsova N, et al. Dapper, a Dishevelled-associated antagonist of beta-catenin and JNK signaling, is required for notochord formation. Dev Cell 2002; 2:449 - 61; http://dx.doi.org/10.1016/S1534-5807(02)00140-5; PMID: 11970895
- Katoh M, Katoh M. Identification and characterization of human DAPPER1 and DAPPER2 genes in silico. Int J Oncol 2003; 22:907 - 13; PMID: 12632086
- Fisher DA, Kivimäe S, Hoshino J, Suriben R, Martin PM, Baxter N, et al. Three Dact gene family members are expressed during embryonic development and in the adult brains of mice. Dev Dyn 2006; 235:2620 - 30; http://dx.doi.org/10.1002/dvdy.20917; PMID: 16881060
- Yau TO, Chan CY, Chan KL, Lee MF, Wong CM, Fan ST, et al. HDPR1, a novel inhibitor of the WNT/beta-catenin signaling, is frequently downregulated in hepatocellular carcinoma: involvement of methylation-mediated gene silencing. Oncogene 2005; 24:1607 - 14; http://dx.doi.org/10.1038/sj.onc.1208340; PMID: 15580286
- Xun WW, Brennan P, Tjonneland A, Vogel U, Overvad K, Kaaks R, et al. Single-nucleotide polymorphisms (5p15.33, 15q25.1, 6p22.1, 6q27 and 7p15.3) and lung cancer survival in the European Prospective Investigation into Cancer and Nutrition (EPIC). Mutagenesis 2011; 26:657 - 66; http://dx.doi.org/10.1093/mutage/ger030; PMID: 21750227
- Li BC, Chan WY, Li CY, Chow C, Ng EK, Chung SC. Allelic loss of chromosome 6q in gastric carcinoma. Diagn Mol Pathol 2003; 12:193 - 200; http://dx.doi.org/10.1097/00019606-200312000-00003; PMID: 14639105
- Carvalho B, van der Veen A, Gärtner F, Carneiro F, Seruca R, Buys CH, et al. Allelic gains and losses in distinct regions of chromosome 6 in gastric carcinoma. Cancer Genet Cytogenet 2001; 131:54 - 9; http://dx.doi.org/10.1016/S0165-4608(01)00514-3; PMID: 11734319
- Qu XY, Hauptschein RS, Rzhetsky A, Scotto L, Chien MC, Ye X, et al. Analysis of a 69-kb contiguous genomic sequence at a putative tumor suppressor gene locus on human chromosome 6q27. DNA Seq 1998; 9:189 - 204; PMID: 10520750
- Rodriguez C, Causse A, Ursule E, Theillet C. At least five regions of imbalance on 6q in breast tumors, combining losses and gains. Genes Chromosomes Cancer 2000; 27:76 - 84; http://dx.doi.org/10.1002/(SICI)1098-2264(200001)27:1<76::AID-GCC10>3.0.CO;2-E; PMID: 10564589
- Minaguchi T, Matsushima M, Saito S, Kanamori Y, Shirahama S, Okamoto S, et al. Complete DNA sequence and characterization of a 330-kb VNTR-rich region on chromosome 6q27 that is commonly deleted in ovarian cancer. DNA Res 1999; 6:131 - 6; http://dx.doi.org/10.1093/dnares/6.2.131; PMID: 10382971
- Liu Y, Emilion G, Mungall AJ, Dunham I, Beck S, Le Meuth-Metzinger VG, et al. Physical and transcript map of the region between D6S264 and D6S149 on chromosome 6q27, the minimal region of allele loss in sporadic epithelial ovarian cancer. Oncogene 2002; 21:387 - 99; http://dx.doi.org/10.1038/sj.onc.1205067; PMID: 11821951
- Wong HC, Bourdelas A, Krauss A, Lee HJ, Shao Y, Wu D, et al. Direct binding of the PDZ domain of Dishevelled to a conserved internal sequence in the C-terminal region of Frizzled. Mol Cell 2003; 12:1251 - 60; http://dx.doi.org/10.1016/S1097-2765(03)00427-1; PMID: 14636582
- Barker N, Clevers H. Mining the Wnt pathway for cancer therapeutics. Nat Rev Drug Discov 2006; 5:997 - 1014; http://dx.doi.org/10.1038/nrd2154; PMID: 17139285
- Yue W, Sun Q, Dacic S, Landreneau RJ, Siegfried JM, Yu J, et al. Downregulation of Dkk3 activates beta-catenin/TCF-4 signaling in lung cancer. Carcinogenesis 2008; 29:84 - 92; http://dx.doi.org/10.1093/carcin/bgm267; PMID: 18048388
- Gao X, Wen J, Zhang L, Li X, Ning Y, Meng A, et al. Dapper1 is a nucleocytoplasmic shuttling protein that negatively modulates Wnt signaling in the nucleus. J Biol Chem 2008; 283:35679 - 88; http://dx.doi.org/10.1074/jbc.M804088200; PMID: 18936100
- Waxman JS, Hocking AM, Stoick CL, Moon RT. Zebrafish Dapper1 and Dapper2 play distinct roles in Wnt-mediated developmental processes. Development 2004; 131:5909 - 21; http://dx.doi.org/10.1242/dev.01520; PMID: 15539487
- Gillhouse M, Wagner Nyholm M, Hikasa H, Sokol SY, Grinblat Y. Two Frodo/Dapper homologs are expressed in the developing brain and mesoderm of zebrafish. Dev Dyn 2004; 230:403 - 9; http://dx.doi.org/10.1002/dvdy.20060; PMID: 15188426
- Waxman JS. Regulation of the early expression patterns of the zebrafish Dishevelled-interacting proteins Dapper1 and Dapper2. Dev Dyn 2005; 233:194 - 200; http://dx.doi.org/10.1002/dvdy.20301; PMID: 15765513
- Teran E, Branscomb AD, Seeling JM. Dpr Acts as a molecular switch, inhibiting Wnt signaling when unphosphorylated, but promoting Wnt signaling when phosphorylated by casein kinase Idelta/epsilon. PLoS One 2009; 4:e5522; http://dx.doi.org/10.1371/journal.pone.0005522; PMID: 19440376
- Kivimäe S, Yang XY, Cheyette BN. All Dact (Dapper/Frodo) scaffold proteins dimerize and exhibit conserved interactions with Vangl, Dvl, and serine/threonine kinases. BMC Biochem 2011; 12:33; http://dx.doi.org/10.1186/1471-2091-12-33; PMID: 21718540
- Su Y, Zhang L, Gao X, Meng F, Wen J, Zhou H, et al. The evolutionally conserved activity of Dapper2 in antagonizing TGF-beta signaling. FASEB J 2007; 21:682 - 90; http://dx.doi.org/10.1096/fj.06-6246com; PMID: 17197390
- Zhang L, Zhou H, Su Y, Sun Z, Zhang H, Zhang L, et al. Zebrafish Dpr2 inhibits mesoderm induction by promoting degradation of nodal receptors. Science 2004; 306:114 - 7; http://dx.doi.org/10.1126/science.1100569; PMID: 15459392
- Jia Y, Yang Y, Brock MV, Zhan Q, Herman JG, Guo M. Epigenetic Regulation of DACT2, A Key Component of the Wnt Signaling Pathway in Human Lung Cancer. [Epub ahead of print] J Pathol 2012; http://dx.doi.org/10.1002/path.4073; PMID: 22806826
- Herman JG, Graff JR, Myöhänen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci U S A 1996; 93:9821 - 6; http://dx.doi.org/10.1073/pnas.93.18.9821; PMID: 8790415
- Zhang W, Glöckner SC, Guo M, Machida EO, Wang DH, Easwaran H, et al. Epigenetic inactivation of the canonical Wnt antagonist SRY-box containing gene 17 in colorectal cancer. Cancer Res 2008; 68:2764 - 72; http://dx.doi.org/10.1158/0008-5472.CAN-07-6349; PMID: 18413743
- Finn RS, Press MF, Dering J, Arbushites M, Koehler M, Oliva C, et al. Estrogen receptor, progesterone receptor, human epidermal growth factor receptor 2 (HER2), and epidermal growth factor receptor expression and benefit from lapatinib in a randomized trial of paclitaxel with lapatinib or placebo as first-line treatment in HER2-negative or unknown metastatic breast cancer. J Clin Oncol 2009; 27:3908 - 15; http://dx.doi.org/10.1200/JCO.2008.18.1925; PMID: 19620495
- Aberle H, Bauer A, Stappert J, Kispert A, Kemler R. beta-catenin is a target for the ubiquitin-proteasome pathway. EMBO J 1997; 16:3797 - 804; http://dx.doi.org/10.1093/emboj/16.13.3797; PMID: 9233789