Abstract
Dietary cadmium exposure was recently found to alter DNA methylation in adults, but data on effects early in life are lacking. Our objective was to evaluate associations between prenatal cadmium exposure, DNA methylation and birth weight. In total 127 mother-child pairs from rural Bangladesh were studied. For comparison, we included 56 children at 4.5 y. Cadmium concentrations in mothers’ blood (gestational week 14) and children’s urine were measured by ICPMS. Global DNA methylation was analyzed by Infinium HumanMethylation450K BeadChip in cord blood and children’s blood. Maternal cadmium exposure was associated with cord blood DNA methylation (p-value < 10–16). The association was markedly sex-specific. In boys, 96% of the top 500 CpG sites showed positive correlations (rS-values > 0.50), whereas most associations in girls were inverse; only 29% were positive (rS > 0.45). In girls we found overrepresentation of methylation changes in genes associated with organ development, morphology and mineralization of bone, whereas changes in boys were found in cell death-related genes. Several individual CpG sites that were positively associated with cadmium were inversely correlated with birth weight, although none statistically significant after correction for multiple comparisons. The associations were, however, fairly robust in multivariable-adjusted linear regression models. We identified CpG sites that were significantly associated with cadmium exposure in both newborns and 4.5-y-old children. In conclusion, cadmium exposure in early life appears to alter DNA methylation differently in girls and boys. This is consistent with previous findings of sex-specific cadmium toxicity. Cadmium-related changes in methylation were also related to lower birth weight.
Introduction
The toxic metal cadmium (Cd) is ubiquitous in the environment. Human exposure occurs mainly via consumption of plant-derived foods, certain types of seafood and offal and via tobacco smoking.Citation1 Chronic health effects of Cd in adults are well documentedCitation2 and there is increasing evidence that also early-life Cd exposure has detrimental effects on child health and development. Cadmium exposure during pregnancy has been associated with decreased birth weight,Citation3,Citation4 which in turn is associated with future disease risk.Citation5,Citation6 In a large longitudinal cohort study, we found a sex-specific association between maternal Cd exposure during pregnancy and infant size at birth, with inverse associations in girls, but little evidence of effects in boys.Citation7 The effects on children’s size seemed to remain until 5 y of age.Citation8
The mechanisms of early-life toxicity of Cd are not yet clear. Suggested mechanisms include disturbed zinc transfer to the fetus,Citation9 interference with glucocorticoid balance,Citation10 and the insulin-like growth factor (IGF) axis,Citation11 all of which may impair fetal growth. A further yet unexplored mechanism of Cd toxicity is interference with the epigenetic machinery, such as DNA methylation, processes that are crucial for early fetal development.Citation12,Citation13 In particular early-life undernutrition have been liked to changes in epigenetic processes, with subsequent consequences for long-term illness.Citation14 In vitro studies have shown Cd-associated gene-specific DNA hypermethylation along with gene silencing, as well as global DNA hypomethylation.Citation15,Citation16 In chick embryos Cd caused downregulation of gene expression of the DNA methyltransferases DNMT3A and 3B.Citation17 In a cross-sectional study of 202 women with low-level environmental Cd exposure from the diet, Cd in urine was associated with DNA hypomethylation of LINE1 retrotransposon sequences, a crude marker for global methylation, in peripheral blood.Citation18 Also, in a study on the role of dietary factors for LINE1 methylation in cord blood, the estimated maternal dietary Cd exposure was associated with LINE1 hypomethylation.Citation19
In this study our aim was to elucidate whether Cd exposure during pregnancy, assessed by individual biomarkers, is associated with altered DNA methylation in the newborn, and in turn, if this may affect the child’s birth weight.
Results
Maternal blood Cd concentrations in gestational week (GW) 14 varied from 0.38 to 5.4 µg/kg (). Blood Cd concentrations in the study population did not differ from those among all women in the cohort, whereas the median Cd in maternal urine (GW8) was slightly higher in this subgroup compared with all women. Blood Cd concentrations in mothers of newborn boys (median 1.2 µg/kg; 5–95th percentile 0.52–3.1) were similar to the Cd concentrations in mothers of newborn girls (1.4 µg/kg; 0.56–3.0). The 4.5-y-old children had lower urinary Cd than the women (Table S1).
Table 1. Characteristics of the 127 mother-child pairs in the present study, as well as all other women who were enrolled in the MINIMat trial from October 2002 through October 2003 and gave live birth (n = 1729)
Cd exposure and DNA methylation in cord blood and 4.5-y-old children
Cd in maternal blood was associated with methylation levels in components 4 and 5 (Figure S1). There was no association with Cd in maternal urine in any of the components and further analyses were therefore performed with Cd in blood only.
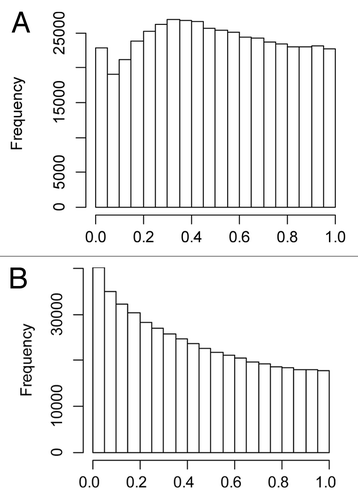
We first evaluated whether the Cd exposure was associated with global DNA methylation by analyzing all CpG sites in cord blood in all newborns in separate models vs. Cd in maternal blood. The analysis showed that small p-values were more frequent than expected from a uniform distribution (Kolmogorov-Smirnov test p-value < 10˗16). There were differences between sexes, in that the effect of Cd exposure seemed to be more pronounced in boys compared with girls ().
Figure 1. The histograms show the frequency distribution of the p-values (x-axis) of the regression coefficients for cadmium from 482, 421 separate regression models, one for each CpG site, of DNA methylation in cord blood vs. cadmium in maternal blood for girls (A) and boys (B), respectively.
We then analyzed CpG-specific effects of Cd. Cadmium vs. DNA methylation showed correlations between rs = -0.36 – 0.43 (range for the top 500 strongest correlations), and the lowest unadjusted p-value was 2.6 × 10−6. After adjusting for multiple comparisons all p-values were > 0.05. The top five genes correlating with Cd () were HISTH4L (rs = 0.43), PAX9 (rs = 0.42), APBB3 (rs = 0.41), GAP43 (rs = 0.41) and PTPRN2 (rs = ˗0.41).
Table 2. Top 10 correlations (rS) between maternal blood Cd concentrations (MB-Cd) and DNA methylation (CpG sites) in cord blood of all infants/newborns as well as for girls and boys separately. Sites are listed according to chromosome number
We then stratified the analysis by sex, as we previously found sex-specific effects of Cd exposure during early life.Citation7 When all probes were considered, there were generally slightly stronger correlations between maternal blood Cd and DNA methylation in cord blood of boys than in girls (, showing the top ten CpG sites for all children and by sex). In boys, the lowest unadjusted p-value observed among all CpG sites was 6.3 x10˗7 (rs = 0.61), whereas in girls it was 0.72 × 10˗7 (rs = ˗0.57), although all p-values adjusted for multiple comparisons were > 0.05. In boys, 96% of the top 500 CpG sites showed positive correlations between maternal Cd concentrations and methylation (all positive β-values had p-values > 0.50), whereas in girls only 29% of the sites showed positive correlations (all positive rs > 0.45) and the remaining were inverse (rs > -0.41).
We performed sex-specific pathway analyses based on annotated genes among the 500 CpG sites with strongest correlations between maternal blood Cd and cord blood methylation. In the top five networks obtained, there were some overlap between boys and girls; e.g., cell morphology, cell cycle, cellular growth and proliferation (Table S2), but they were different in ranking. Sex-specific differences were found for specific functions. Girls showed the strongest associations between Cd and DNA methylation in CpG sites related to embryonic and organ development, especially connective tissue and skeletal development (bone mineralization and morphology of the bone; altogether around 40 genes; p = 1.5×10˗5). Boys, by contrast, showed the strongest associations between Cd and DNA methylation in genes involved in cell death (105 genes among the top 500; p-value 3.9×10˗6). As mentioned above, CpG sites in those genes showed more methylation in boys with increasing maternal blood Cd concentrations.
Among the 500 top CpG sites in cord blood (both sexes) that showed the strongest correlations between methylation and Cd in maternal blood, we selected those that were also significantly correlated in the same direction in the 4.5-y-old children (child blood CpG methylation vs. child urinary Cd; ). Cadmium exposure was inversely correlated with methylation in the CpG site cg16001202, close to SMOC2, in both cord blood and children’s blood, and the same direction of correlation, but non-significant, was found for two more sites in SMOC2 (not shown in ). Similarly, one CpG site each in IL17RD, C4B, RAB23, MYPN, NODAL, PLCG2 and NAA10 and one CpG-site each on chromosomes 8 (closest gene RUNX1T1), 10 (CASC2/ FAM204A), and 13 in both cord blood and children’s blood were significantly correlated in the same direction with Cd exposure. The function of these genes is shown in Supplemental Table S3 and some examples are shown in . There was no indication of sex-related differences between Cd and DNA methylation for these sites (data not shown). The effect estimates or the strength of associations did not change substantially after adjustments of other influential variables (). After adjustments, the associations for C4B, RUNX1T1, MYPN and the CpG site on chromosome 13 remained significant in 4.5-y-old children, and ILI7RD, SMOC2, NODAL, PLCG2 were close to significant. We additionally adjusted for betel chewing and food and micronutrient supplementation in a separate analysis but it did not change the results (data not shown).
Table 3. CpG sites that showed significant correlations (rS) in the same direction both between Cd in maternal blood (MB-Cd) and DNA methylation in cord blood and between Cd in urine (U-Cd) and DNA methylation in blood from 4.5-y-old children
Figure 2. Scatterplots depicting (A) fraction of DNA methylation in cord blood for the CpG site cg20309121 in RUNX1T1 vs. maternal blood Cd at GW14; and (B) fraction of DNA methylation for cg20309121 in peripheral blood from 4.5-y-old children vs. their urinary Cd; (C) fraction of DNA methylation in cord blood for cg09127607 in MYPN vs. maternal blood Cd at GW14; and (D) fraction of DNA methylation for cg09127607 in peripheral blood from 4.5-y-old children vs. their urinary Cd. Solid lines represent Lowess-moving average curves; dashed lines represent fitted curves from the multivariable-adjusted regression analyses defined in .
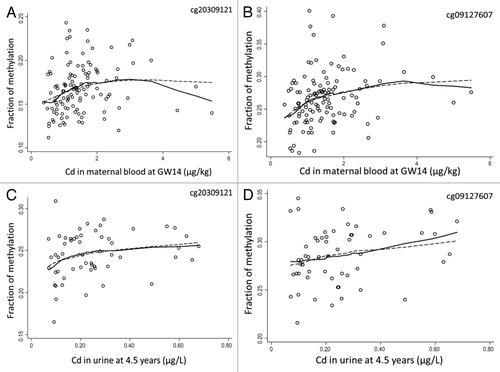
In general, the effect of Cd on degree of CpG methylation ranged between 0.5–1.4% for a doubling of blood Cd (µg/kg) in the mothers (based on significant effect estimates in both cord blood and 4.5-y-old children in ).
Maternal Cd, DNA methylation and birth weight
Among the top 500 CpG sites in cord blood with strongest correlation between methylation and Cd in maternal blood, multiple sites also correlated significantly to birth weight (; unadjusted p-values shown, none of which remained statistically significant when adjusted for multiple comparisons). We focused on relationships where: (1) DNA methylation was positively correlated with blood Cd and inversely correlated with birth weight, or (2) DNA methylation was inversely correlated with blood Cd and positively correlated with birth weight (). All associations found when considering both girls and boys were of type 1. We then stratified for sex and found for girls two CpG sites of interest: TSH7DA (cg07846874) on chromosome 7 and one site on chromosome X (cg11595135) that were positively associated with Cd (rs = 0.51, unadjusted p = 0.00007 and rs = 0.44, p = 0.00082, respectively) and inversely associated with birth weight (rs = -0.35, p = 0.005; and rs = -0.37, p = 0.003, respectively). In girls, we also found one site on chromosome 2 (cg00224807) that was inversely associated with maternal blood Cd (rs = -0.43, p = 0.0012) and positively associated with birth weight (rs = 0.26, p = 0.036). For boys, we found one site on chromosome 3 (cg19119945) that was inversely associated with maternal blood Cd (rs = -0.48, p = 0.00016) and positively associated with birth weight (rs = 0.27, p = 0.036).
Table 4. CpG sites in cord blood that were significantly correlated (rS) to both to Cd in maternal blood (MB-Cd) and birth weight
The multivariable-adjusted linear regression analyses for all newborns showed that the associations between Cd-related DNA methylation and birth weight were fairly robust (), and CpG sites in PTTG1, THSD7A, BCCIP, TMEM179, SRP14 and GDPD1 remained statistically significant after adjustment. We also adjusted for food and micronutrient supplementation (Table S4) and betel chewing in a separate analysis but it did not substantially change the results. For the CpG sites presented in –, only three out of 63 sites contained a SNP within ≤ 10bp from the query site (Table S5). One of these SNPs has a minor allele frequency of 0.017, for the other two SNPs no allele frequencies were available. Ten out of 63 CpG sites showed a SNP > 10 bp from query site, but for four of these SNPs no allele frequencies were available or the minor allele frequency was very low.
Discussion
This study indicates that low-level environmental Cd exposure during early pregnancy is associated with sex-specific alterations in DNA methylation in fetal blood. On a global level the effect of Cd was most evident in boys. Boys showed generally global hypermethylation. In contrast, the newborn girls showed markedly more hypomethylation. Interestingly, hypomethylation of repetitive sequences, a crude marker for global methylation, in relation to environmental Cd exposure has been reported for adult women.Citation18 Moreover, in rats exposed to Cd during pregnancy, CpG sites in the hepatic glucocorticoid receptor were hypomethylated in relation to Cd in female fetuses but hypermethylated in male fetuses.Citation20 The sex differences in methylation in relation to Cd exposure were further highlighted in the analysis of specific gene functions and pathways; altered methylation of genes for bone morphology and mineralization was found in cord blood of girls, whereas hypermethylation of genes involved in cell death was found in boys. This is noteworthy in relation to the previous findings of the inverse associations of Cd exposure with fetal size, in particular size of the head and femur length, in girls, but not in boys.Citation21 Also, Cd is associated with osteoporosis and fractures particularly in women.Citation22 The observed sex-difference is noteworthy considering the emerging data on obvious sexual dimorphism in environmental epigenetic programming.Citation14
We found methylation changes in CpG sites in genes associated with both Cd and birth weight when considering all children. The data could not clearly support our hypothesis that girls preferentially should have Cd-related methylation in genes regulating infant size. However, this might have been a question of low statistical power. The multivariable-adjusted analyses indicated a fairly large impact of DNA methylation in the selected sites on birth weight, for one percent-unit increment in DNA methylation; this would correspond to a decrease in birth weight of 7 g to 46 g. This emphasizes the notion that CpG methylation is very important for fetal development. Each doubling of µg/kg Cd in maternal blood resulted in an approximately 1% change in DNA methylation, which theoretically would correspond to up to a 46 g reduction in birth weight. This Cd-related reduction in birth weight is similar in size to what has been observed in girls, a reduction of 45 g per every 1 µg/L increment of Cd in maternal urine.Citation7
We found similar Cd-related methylation changes in several specific CpG sites in both cord blood and blood of 4.5-y-old children. These findings suggest that some specific Cd-related changes persist at least to later in childhood, and may be important for later health effects. Thus, it is essential to follow these children to evaluate if the changes become modified later in life.
Some methodological aspects need to be commented upon. It is clear from our study that children’s sex was influential for the Cd-related changes. Stratifying for sex markedly increased the effect sizes, largely because of the different directions of the Cd-related DNA methylation in boys and girls. The phenomenon of sex-specific effects on DNA methylation is not limited to exposure to Cd, but occurs for other exposures as well.Citation23 Sex-specific DNA methylation has been reported in whole blood and saliva samples from adults.Citation24 One can speculate that Cd might interfere with already existing sex-specific gene expression, or alternatively it causes sex-specific DNA methylation for genes that should not differ between the sexes. However, to our knowledge there is no study that has characterized sex-specific gene expression per se in cord blood to compare with our data. When comparing the sex-specific genes identified in the study with adults by Liu et al.Citation24 with our data, we found no overlap with the top genes for associations between Cd and DNA methylation that we report in our study.
We measured DNA methylation in cord blood mononuclear cells, which is a mixture of different cell types with partly different methylation patterns, in particular for cell type-specific immune functionsCitation25 that may blur associations between DNA methylation and Cd exposure. We were not able to sort cells in the blood samples during the field studies. Houseman and coworkersCitation26 describe a method for inferring changes in the distribution of white blood cells between different subpopulations (e.g., cases and controls) using DNA methylation signatures. We did not apply this method since there were no clearly identified subpopulations in our study population (i.e., exposed and non-exposed newborns). Further, the DNA methylation signatures developed by Houseman et al.Citation26 were based on whole blood from different adult populations, which are not directly applicable here as they probably differ from cord blood. To note, Cd exposure is not associated with major changes in the blood cell profile, and although subtle changes cannot be ruled out, we consider it unlikely that our results represent shifts in leukocyte profile from Cd exposure. The consistency of several associations in cord blood and in 4.5-y-old children support that the findings indeed were true, and persist during early child development.
It should be noted that none of the correlations for specific CpG sites were statistically significant after adjustments for multiple comparisons and the specific genes identified here must be verified in new studies. Low power is a general problem when analyzing – omics data. Nevertheless, in the multivariable-adjusted linear regression analyses most associations were robust, also when taking other influential factors into account. For the pathway analysis the IPA software uses Fisher’s exact test, which does not consider gene dependence structure and pathway hierarchical dependence structures. This might lead to too optimistic results.
Cadmium concentrations in maternal blood were more strongly associated with DNA methylation in cord blood than Cd in urine, which is reasonable, as the blood Cd to a larger extent represents the ongoing exposure, whereas Cd in urine represents the maternal body burden, mainly the accumulation in liver and kidneys.Citation2 We did not measure Cd in blood for the 4.5-y-old children, but in urine, which probably reflects, to a larger extent, ongoing Cd exposure when they are young, and thus, have not yet accumulated much Cd in their kidneys.
To our knowledge, none of the genes linked to CpG sites presented here have previously been associated with Cd exposure or Cd toxicity and the results need to be cautiously interpreted. One exception is the PTTG1, which was more expressed in liver cancer cells in response to Cd exposure in vitro.Citation27 In contrast, in the present study on healthy newborns PTTG1 was more methylated with increasing Cd exposure. Methylation in C4B, RUNX1T1 and MYPN was significantly associated with Cd-exposure both in newborns and in children at 4.5 y. C4B encodes the basic form of complement factor 4, part of the classical complement activation pathway. RUNX1T1 is the gene for runt-related transcription factor 1; translocated to, 1 (cyclin D-related), a transcription factor that regulates critical processes in many aspects of hematopoiesis.Citation28 MYPN encodes myopalladin, a protein that regulates the formation of striated muscles in vertebrates.Citation29 In contrast to these three genes, few of the other genes that we found to be significantly associated with both Cd and birth weight have previously been related to developmental growth. BCCIP is important for structural stability of chromosomes and conditional BCCIP knock-down transgenic mice show growth retardation and impaired embryonic cell proliferation.Citation30,Citation31 Thus, one could speculate that in humans increased methylation of BCCIP results in reduced growth of the developing child. To note, one CpG site was located in the HOX cluster, between HOXC12 and HOXC13, on the long arm chromosome 12. The homeobox (HOX) family encodes transcription factors that serve as regulators in initiating developmental programs.Citation32 THSD7A was associated with birth weight both in the analyses for all children and for just girls. THSD7A is a protein described in zebrafish to be involved in angiogenesis by endothelial migration.Citation33
In conclusion, Cd exposure in pregnancy alters fetal DNA methylation in a sex-specific manner. This may explain previous findings of differences in toxicity of Cd between girls and boys. Some Cd-related DNA methylation changes were also related to lower birth weight, but the consequences for child health and development remains to be elucidated.
Materials and methods
Study area and subjects
Our studies on early-life Cd effects on fetal and child health and development were nested into a randomized population-based food and micronutrient supplementation trial in pregnancy (MINIMat trial)Citation34 involving 4436 pregnant women recruited from November 2001 through October 2003. Study area was the Matlab sub-district located approximately 50 km southeast of Dhaka, Bangladesh, where the International Centre for Diarrheal Disease Research, Bangladesh (icddr,b) runs a health and demographic surveillance system (HDSS), a hospital and four health clinics.
Pregnancy was initially identified by urine test, which was offered to women who reported that their last menstrual period (LMP) was overdue at the monthly home visit by community health research workers. In case of a positive result, the women were invited to participate in the MINIMat trial. The eligibility criteria for enrollment in the MINIMat trial included viable fetus, gestational age < 14 weeks, no severe illness, and consent for participation.Citation34 The intervention included randomized supplementation of both food [early invitation at GW9 or usual invitation at approximately GW20] and micronutrients (two groups with different combinations of iron and folic acid, one with 13 additional micronutrients), resulting in six different groups.Citation34,Citation35
For the present study of Cd exposure and DNA methylation we used a sub-sample of 127 women who were enrolled from October 2002 through October 2003, gave singleton birth at the health care facilities during early day-time, and had cord blood collected at delivery.Citation36 The main reasons for the low number of deliveries was the high frequency of home deliveries (> 60%)Citation37 and that many deliveries at the health facilities occurred in late afternoon or night, when the logistics didn’t allow for processing and transporting of samples to the laboratory in Dhaka. In comparison to all other women who were enrolled in the MINIMat trial from October 2002 through October 2003 and gave singleton birth (n = 1729), these 127 women had slightly higher SES and were more likely to be primiparous ().
For comparison, we also studied Cd-related DNA methylation in blood mononuclear cells from 56 children at 4.5 y of age (Table S1). These children also originated from the MINIMat trial, but they were independent of the 127 pregnant women and were therefore used as an independent comparison group for follow-up of Cd-related DNA methylation later in life.
Participants gave written, informed consent, and the study was approved by the ethical review committees at icddr,b, in Bangladesh and at Karolinska Institutet, Sweden.
Exposure assessment
We measured Cd in maternal blood (erythrocyte fraction; hereafter referred to as blood Cd), a marker of ongoing exposure. We also measured Cd in maternal and child urine, which reflects long-term exposure. Maternal blood and urine samples were collected at GW8 and 14, respectively. Sample collection was performed either at the health care facilities or at home. Urine was collected in acid-washed plastic vials and blood in 5.5 mL Li-Heparin tubes (Germany).
Cadmium (Cd111) in urine and blood was measured with inductively coupled plasma mass spectrometry (ICPMS; model 7500ce; Agilent Technologies, Tokyo, Japan) at Karolinska Institutet (Stockholm, Sweden) with correction cell in helium mode. The sample preparation and details concerning the ICPMS analyses have been described in detail elsewhere.Citation38 No samples were below limit of detection (overall < 0.01 μg/L) and the quality control showed good agreement with the recommended concentrations. To compensate for variation in urine dilution, we adjusted for the average specific gravity of the urine (1.012 g/mL both in maternal and child urine).
DNA isolation and epigenetic analysis
DNA was isolated using QIAamp DNA Blood Mini kit (Qiagen) at icddr,b (Dhaka, Bangladesh). DNA quality was evaluated on a NanoDrop spectrophotometer (NanoDrop Products, Wilmington, DE, USA) and a Bioanalyzer 2100 (Agilent, Santa Clara, CA, USA) and showed good quality (260/280 nm > 1.80). One µg DNA (50 ng/µL) was bisulfite-treated using the EZ DNA Methylation kit (Zymo, D5001). Cord blood DNA samples were randomized for sex and maternal blood Cd concentrations on two 96-well plates for epigenetic analysis with the Infinium HumanMethylation450K BeadChip (lllumina, San Diego, CA, USA). The samples from 4.5 y-old children were all positioned in one of the 96-well plates, but randomized for sex and Cd exposure within the plate. We also included four controls in duplicate, with each duplicate positioned on different 96-well plates and on different HumanMethylation450K BeadChips: three control samples were DNA extracted from blood and one sample was demethylated DNA (from Zymo). The SCIBLU facility (Lund, Sweden) used 200 ng bisulphite-treated DNA, diluted with H20 to 4 µl, for hybridization to the Infinium HumanMethylation450K BeadChip following the manufacturer’s instructions. Unsupervised hierarchical cluster analysis showed that all duplicate clustered together despite being on different plates.
Birth weight and covariates
Birth weight was measured mostly within 24 h of delivery, using electronic scales (SECA pediatric scales, Hamburg, Germany) with precision of 10 g.Citation37 Gestational age was calculated by subtracting the date of LMP from the date of delivery. After delivery, mothers were asked about their smoking and betel chewing habits during pregnancy. None of the women reported smoking, whereas 58% reported betel chewing. For assessment of socioeconomic status (SES) an asset index, based mainly on house construction and household assets, was calculated using principal component analysis (PCA).Citation39 Height and weight of the children at 4.5 y of age were converted to age- and sex-standardized z-scores [weight-for-age (WAZ), and height-for-age (HAZ)], using the World Health Organization growth references].Citation40
Because we have previously shown that the present study population is exposed to arsenic (As) via drinking waterCitation41,Citation42 and As exposure has been associated with DNA methylation,Citation43 we adjusted for concentrations of urinary As in the statistical models. Measurements of inorganic As and its methylated metabolites had previously been measured in maternal urine during pregnancy (GW8; hereafter referred to as As in urine) by hydride generation atomic absorption spectrometry,Citation42 and in children’s urine at 4.5 y of age by high-performance liquid chromatography online with hydride generation and ICPMS.Citation41
Statistical analysis
Methylation levels are specified by so-called β-values, which represent the fraction of methylation and hence range from 0 (unmethylated) to 1 (fully methylated). Beta-values were extracted from BeadStudio software (lllumina, San Diego, CA, USA). Missing values (0.1%) were imputed using k-nearest neighbor imputation (k = 10).
Principal component analysis (PCA) captures the major directions of variation in the data. The first principal component accounts for as much of the variability in the data as possible, and each succeeding component in turn accounts for as much of the variability possible given it is uncorrelated with the preceding components. For PCA, the R package ‘swamp’ was employed. For each of the top principal components we fitted a univariate linear model with each of the sample annotations as regressor. The log10 p-values of the models’ F-statistics were plotted as a heat map (Figure S1). PCA was run separately for newborns and 4.5-y-old children. The analysis for cord blood samples showed that analysis plate (two 96-well plates) was associated with cord methylation levels in the first (p = 10˗6) and third components (p < 10˗10). We removed the plate influence at each CpG site by using the residuals from the initial linear regression model of methylation with analysis plate as regressor. The residuals of the linear model added to the total mean before correction became the new data for each CpG site. In this way, the levels of each of the methylation sites were unrelated to analysis plate. No other variables had a major impact on general DNA methylation.
We evaluated whether the Cd exposure was associated with global DNA methylation by performing 482,421 separate linear regression models, one for each CpG site, where CpG methylation was the dependent variable and Cd concentrations the only independent variable. The range of Cd was rescaled to vary between 0 and 1. We tested whether the slope was statistically significant in all 482,421 models; if there was no effect of Cd on any CpG site, then the slopes would result from mere sampling error. In this case the p-values would be distributed uniformly over the 0-to-1 range.
The associations of specific CpG site methylation levels with variables of interest were then evaluated by Spearman correlation. Resulting p-values were corrected for multiple testing (n = 482,421) by the Benjamini-Hochberg method to obtain false discovery rates (FDR). Among the 500 top correlations of Cd in maternal blood with methylation in CpG sites in all newborns, we further analyzed those that also were correlated with birth weight. In addition, we compared the correlations to those in 4.5-y-old children. The associations with methylation in the CpG sites were subsequently evaluated using scatterplots and multivariable-adjusted regression modeling.
In general, associations of DNA methylation in different CpG sites with Cd exposure (maternal blood Cd and children’s urinary Cd) and birth weight showed linear patterns, hence linear regression analyses were applied. Because the Cd concentrations in maternal blood and children’s urine were left skewed () these variables were log2-transformed. We did not observed any genotype-related clustering of DNA methylation when visually inspecting associations between Cd and DNA methylation, and associations between birth weight and DNA methylation. When evaluating DNA methylation in cord blood in relation to maternal blood Cd concentrations, we adjusted for maternal age, BMI (GW8), SES, gestational age at birth, fetal sex and maternal urinary As (GW8). A similar strategy was applied when children’s urinary Cd (4.5 y) was used as an exposure marker. We adjusted the model by children’s concurrent age, HAZ, SES, children’s concurrent urinary As, and sex of the child. When assessing the association between DNA methylation in cord blood and birth weight, we adjusted for maternal age, BMI (GW8), SES, gestational age at birth, fetal sex, maternal blood Cd and maternal urinary As. To evaluate potential sex differences, we also stratified the analyses by sex. In sensitivity analyses we tested if betel chewing or the food and micronutrient supplementation ingested during pregnancy had any impact on the above-mentioned associations.
For the bioinformatics analysis we used the Ingenuity Pathway Analysis Tool (IPA Tool; Ingenuity H Systems; Redwood City, CA, USA; www.ingenuity.com). We selected the top 500 CpG sites associated with Cd in maternal blood: that resulted in 390 gene-annotated (obtained from Illumina) CpG sites in boys, 348 annotated CpG sites in girls. In IPA, differentially methylated genes are mapped to genetic networks available in the Ingenuity database and then ranked by score. The Ingenuity Pathway Knowledge Base is derived from known functions and interactions of genes in the literature. The networks created are ranked depending on the number of significantly methylated genes they contain and also list diseases that were most significant.
| Abbreviations: | ||
| As | = | arsenic |
| BMI | = | body mass index |
| Cd | = | cadmium |
| CHR | = | chromosome |
| GW | = | gestational week |
| HDSS | = | health demographic surveillance system |
| HAZ | = | height-for-age z-score |
| inductively coupled plasma mass spectrometry | = | ICPMS |
| LMP | = | last menstrual period |
| MB-Cd | = | maternal blood cadmium |
| PCA | = | principal component analysis |
| SES | = | socioeconomic status |
| WAZ | = | weight-for-age z-score |
| U-As | = | urinary arsenic metabolites |
| U-Cd | = | urinary cadmium |
Additional material
Download Zip (294 KB)Acknowledgments
This research was supported by the Swedish Council for Working Life and Social Research, Karolinska Institutet, Kungliga fysiografiska sällskapet, the European Union (FP6; “PHIME” FOOD-CT-2006–016253) and Sida (contribution no. 75000431, 75000512, 82030018) at the International Centre for Diarrheal Disease Research, Bangladesh (icddr,b).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Olsson IM, Bensryd I, Lundh T, Ottosson H, Skerfving S, Oskarsson A. Cadmium in blood and urine--impact of sex, age, dietary intake, iron status, and former smoking--association of renal effects. Environ Health Perspect 2002; 110:1185 - 90; http://dx.doi.org/10.1289/ehp.021101185; PMID: 12460796
- Järup L, Åkesson A. Current status of cadmium as an environmental health problem. Toxicol Appl Pharmacol 2009; 238:201 - 8; http://dx.doi.org/10.1016/j.taap.2009.04.020; PMID: 19409405
- Salpietro CD, Gangemi S, Minciullo PL, Briuglia S, Merlino MV, Stelitano A, et al. Cadmium concentration in maternal and cord blood and infant birth weight: a study on healthy non-smoking women. J Perinat Med 2002; 30:395 - 9; http://dx.doi.org/10.1515/JPM.2002.061; PMID: 12442603
- Shirai S, Suzuki Y, Yoshinaga J, Mizumoto Y. Maternal exposure to low-level heavy metals during pregnancy and birth size. J Environ Sci Health A Tox Hazard Subst Environ Eng 2010; 45:1468 - 74; http://dx.doi.org/10.1080/10934529.2010.500942; PMID: 20694885
- Barker DJ. Developmental origins of adult health and disease. J Epidemiol Community Health 2004; 58:114 - 5; http://dx.doi.org/10.1136/jech.58.2.114; PMID: 14729887
- Kerkhof GF, Leunissen RW, Hokken-Koelega AC. Early origins of the metabolic syndrome: role of small size at birth, early postnatal weight gain, and adult IGF-I. J Clin Endocrinol Metab 2012; 97:2637 - 43; http://dx.doi.org/10.1210/jc.2012-1426; PMID: 22564668
- Kippler M, Tofail F, Gardner R, Rahman A, Hamadani JD, Bottai M, et al. Maternal cadmium exposure during pregnancy and size at birth: a prospective cohort study. Environ Health Perspect 2011; 120:284 - 9; http://dx.doi.org/10.1289/ehp.1103711; PMID: 21862444
- Gardner R, Kippler M, Tofail F, Bottai M, Hamadani J, Grander M, et al. Environmental exposures to metals and children’s growth to five years: a prospective cohort study. Am J Epidemiol 2013; Accepted
- Kippler M, Hoque AM, Raqib R, Öhrvik H, Ekström EC, Vahter M. Accumulation of cadmium in human placenta interacts with the transport of micronutrients to the fetus. Toxicol Lett 2010; 192:162 - 8; http://dx.doi.org/10.1016/j.toxlet.2009.10.018; PMID: 19854248
- Yang K, Julan L, Rubio F, Sharma A, Guan H. Cadmium reduces 11 beta-hydroxysteroid dehydrogenase type 2 activity and expression in human placental trophoblast cells. Am J Physiol Endocrinol Metab 2005; 290:E135 - 42; http://dx.doi.org/10.1152/ajpendo.00356.2005; PMID: 16144812
- Turgut S, Kaptanoglu B, Turgut G, Emmungil G, Genç O. Effects of cadmium and zinc on plasma levels of growth hormone, insulin-like growth factor I, and insulin-like growth factor-binding protein 3. Biol Trace Elem Res 2005; 108:197 - 204; http://dx.doi.org/10.1385/BTER:108:1-3:197; PMID: 16327072
- Reik W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature 2007; 447:425 - 32; http://dx.doi.org/10.1038/nature05918; PMID: 17522676
- Smith ZD, Chan MM, Mikkelsen TS, Gu H, Gnirke A, Regev A, et al. A unique regulatory phase of DNA methylation in the early mammalian embryo. Nature 2012; 484:339 - 44; http://dx.doi.org/10.1038/nature10960; PMID: 22456710
- Gabory A, Attig L, Junien C. Developmental programming and epigenetics. Am J Clin Nutr 2011; 94:Suppl 1943S - 52S; http://dx.doi.org/10.3945/ajcn.110.000927; PMID: 22049164
- Benbrahim-Tallaa L, Waterland RA, Dill AL, Webber MM, Waalkes MP. Tumor suppressor gene inactivation during cadmium-induced malignant transformation of human prostate cells correlates with overexpression of de novo DNA methyltransferase. Environ Health Perspect 2007; 115:1454 - 9; PMID: 17938735
- Jiang G, Xu L, Song S, Zhu C, Wu Q, Zhang L, et al. Effects of long-term low-dose cadmium exposure on genomic DNA methylation in human embryo lung fibroblast cells. Toxicology 2008; 244:49 - 55; http://dx.doi.org/10.1016/j.tox.2007.10.028; PMID: 18077075
- Doi T, Puri P, McCann A, Bannigan J, Thompson J. Epigenetic effect of cadmium on global de novo DNA hypomethylation in the cadmium-induced ventral body wall defect (VBWD) in the chick model. Toxicol Sci 2011; 120:475 - 80; http://dx.doi.org/10.1093/toxsci/kfr022; PMID: 21278052
- Hossain MB, Vahter M, Concha G, Broberg K. Low-Level Environmental Cadmium Exposure is Associated with DNA Hypomethylation in Argentinean Women. Environ Health Perspect 2012; 120:879 - 84; http://dx.doi.org/10.1289/ehp.1104600; PMID: 22398305
- Boeke CE, Baccarelli A, Kleinman KP, Burris HH, Litonjua AA, Rifas-Shiman SL, et al. Gestational intake of methyl donors and global LINE-1 DNA methylation in maternal and cord blood: prospective results from a folate-replete population. Epigenetics 2012; 7:253 - 60; http://dx.doi.org/10.4161/epi.7.3.19082; PMID: 22430801
- Castillo P, Ibáñez F, Guajardo A, Llanos MN, Ronco AM. Impact of cadmium exposure during pregnancy on hepatic glucocorticoid receptor methylation and expression in rat fetus. PLoS ONE 2012; 7:e44139; http://dx.doi.org/10.1371/journal.pone.0044139; PMID: 22957049
- Kippler M, Wagatsuma Y, Rahman A, Nermell B, Persson LÅ, Raqib R, et al. Environmental exposure to arsenic and cadmium during pregnancy and fetal size: a longitudinal study in rural Bangladesh. Reprod Toxicol 2012; 34:504 - 11; http://dx.doi.org/10.1016/j.reprotox.2012.08.002; PMID: 22985739
- Engström A, Michaëlsson K, Vahter M, Julin B, Wolk A, Åkesson A. Associations between dietary cadmium exposure and bone mineral density and risk of osteoporosis and fractures among women. Bone 2012; 50:1372 - 8; http://dx.doi.org/10.1016/j.bone.2012.03.018; PMID: 22465267
- Hochstenbach K, van Leeuwen DM, Gmuender H, Gottschalk RW, Løvik M, Granum B, et al. Global gene expression analysis in cord blood reveals gender-specific differences in response to carcinogenic exposure in utero. Cancer Epidemiol Biomarkers Prev 2012; 21:1756 - 67; http://dx.doi.org/10.1158/1055-9965.EPI-12-0304; PMID: 22879202
- Liu J, Morgan M, Hutchison K, Calhoun VD. A study of the influence of sex on genome wide methylation. PLoS One 2010; 5:e10028; http://dx.doi.org/10.1371/journal.pone.0010028; PMID: 20386599
- Reinius LE, Acevedo N, Joerink M, Pershagen G, Dahlén SE, Greco D, et al. Differential DNA methylation in purified human blood cells: implications for cell lineage and studies on disease susceptibility. PLoS ONE 2012; 7:e41361; http://dx.doi.org/10.1371/journal.pone.0041361; PMID: 22848472
- Houseman EA, Accomando WP, Koestler DC, Christensen BC, Marsit CJ, Nelson HH, et al. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics 2012; 13:86; http://dx.doi.org/10.1186/1471-2105-13-86; PMID: 22568884
- Kawata K, Shimazaki R, Okabe S. Comparison of gene expression profiles in HepG2 cells exposed to arsenic, cadmium, nickel, and three model carcinogens for investigating the mechanisms of metal carcinogenesis. Environ Mol Mutagen 2009; 50:46 - 59; http://dx.doi.org/10.1002/em.20438; PMID: 19031421
- Lam K, Zhang DE. RUNX1 and RUNX1-ETO: roles in hematopoiesis and leukemogenesis. Front Biosci 2012; 17:1120 - 39; http://dx.doi.org/10.2741/3977; PMID: 22201794
- Bang ML, Mudry RE, McElhinny AS, Trombitás K, Geach AJ, Yamasaki R, et al. Myopalladin, a novel 145-kilodalton sarcomeric protein with multiple roles in Z-disc and I-band protein assemblies. J Cell Biol 2001; 153:413 - 28; http://dx.doi.org/10.1083/jcb.153.2.413; PMID: 11309420
- Huang YY, Lu H, Liu S, Droz-Rosario R, Shen Z. Requirement of mouse BCCIP for neural development and progenitor proliferation. PLoS ONE 2012; 7:e30638; http://dx.doi.org/10.1371/journal.pone.0030638; PMID: 22292003
- Lu H, Huang YY, Mehrotra S, Droz-Rosario R, Liu J, Bhaumik M, et al. Essential roles of BCCIP in mouse embryonic development and structural stability of chromosomes. PLoS Genet 2011; 7:e1002291; http://dx.doi.org/10.1371/journal.pgen.1002291; PMID: 21966279
- Pick L, Heffer A. Hox gene evolution: multiple mechanisms contributing to evolutionary novelties. Ann N Y Acad Sci 2012; 1256:15 - 32; http://dx.doi.org/10.1111/j.1749-6632.2011.06385.x; PMID: 22320178
- Wang CH, Chen IH, Kuo MW, Su PT, Lai ZY, Wang CH, et al. Zebrafish Thsd7a is a neural protein required for angiogenic patterning during development. Dev Dyn 2011; 240:1412 - 21; http://dx.doi.org/10.1002/dvdy.22641; PMID: 21520329
- Persson LÅ, Arifeen S, Ekström EC, Rasmussen KM, Frongillo EA, Yunus M, MINIMat Study Team. Effects of prenatal micronutrient and early food supplementation on maternal hemoglobin, birth weight, and infant mortality among children in Bangladesh: the MINIMat randomized trial. JAMA 2012; 307:2050 - 9; http://dx.doi.org/10.1001/jama.2012.4061; PMID: 22665104
- Tofail F, Persson LÅ, El Arifeen S, Hamadani JD, Mehrin F, Ridout D, et al. Effects of prenatal food and micronutrient supplementation on infant development: a randomized trial from the Maternal and Infant Nutrition Interventions, Matlab (MINIMat) study. Am J Clin Nutr 2008; 87:704 - 11; PMID: 18326610
- Ahmed S, Ahsan KB, Kippler M, Mily A, Wagatsuma Y, Hoque AM, et al. In utero arsenic exposure is associated with impaired thymic function in newborns possibly via oxidative stress and apoptosis. Toxicol Sci 2012; 129:305 - 14; http://dx.doi.org/10.1093/toxsci/kfs202; PMID: 22713597
- Rahman A, Vahter M, Smith AH, Nermell B, Yunus M, El Arifeen S, et al. Arsenic exposure during pregnancy and size at birth: a prospective cohort study in Bangladesh. Am J Epidemiol 2008; 169:304 - 12; http://dx.doi.org/10.1093/aje/kwn332; PMID: 19037006
- Kippler M, Lönnerdal B, Goessler W, Ekström EC, Arifeen SE, Vahter M. Cadmium interacts with the transport of essential micronutrients in the mammary gland - a study in rural Bangladeshi women. Toxicology 2009; 257:64 - 9; http://dx.doi.org/10.1016/j.tox.2008.12.009; PMID: 19126424
- Gwatkin DR, Rustein S, Pande J, Wagstaff RP. Socio-economic differences i health, nutrition, and population in Bangladesh. Washington 2000.
- de Onis M, Onyango AW, Borghi E, Siyam A, Nishida C, Siekmann J. Development of a WHO growth reference for school-aged children and adolescents. Bull World Health Organ 2007; 85:660 - 7; http://dx.doi.org/10.2471/BLT.07.043497; PMID: 18026621
- Gardner R, Hamadani J, Grandér M, Tofail F, Nermell B, Palm B, et al. Persistent exposure to arsenic via drinking water in rural Bangladesh despite major mitigation efforts. Am J Public Health 2011; 101:Suppl 1 S333 - 8; http://dx.doi.org/10.2105/AJPH.2010.300025; PMID: 21778503
- Vahter ME, Li L, Nermell B, Rahman A, El Arifeen S, Rahman M, et al. Arsenic exposure in pregnancy: a population-based study in Matlab, Bangladesh. J Health Popul Nutr 2006; 24:236 - 45; PMID: 17195565
- Ren X, McHale CM, Skibola CF, Smith AH, Smith MT, Zhang L. An emerging role for epigenetic dysregulation in arsenic toxicity and carcinogenesis. Environ Health Perspect 2010; 119:11 - 9; http://dx.doi.org/10.1289/ehp.1002114; PMID: 20682481