Abstract
Lysine methylation of histones and non-histone proteins has emerged in recent years as a posttranslational modification with wide-ranging cellular implications beyond epigenetic regulation. The molecular interactions between lysine methyltransferases and their substrates appear to be regulated by posttranslational modifications surrounding the lysine methyl acceptor. Two very interesting examples of this cross-talk between methyl-lysine sites are found in the SET (Su(var)3–9, Enhancer-of-zeste, Trithorax) domain-containing lysine methyltransferases SET7 and SETDB1, whereby the histone H3 trimethylated on lysine 4 (H3K4me3) modification prevents methylation by SETDB1 on H3 lysine 9 (H3K9) and the histone H3 trimethylated on lysine 9 (H3K9me3) modification prevents methylation by SET7 on H3K4. A similar cross-talk between posttranslational modifications regulates the functions of non-histone proteins such as the tumor suppressor p53 and the DNA methyltransferase DNMT1. Herein, in cis effects of acetylation, phosphorylation, as well as arginine and lysine methylation on lysine methylation events will be discussed.
Introduction
The genome of eukaryotic organisms is laid down on a proteinaceous foundation, the histone octamer, and wrapped around it to form the basic unit of chromatin, the nucleosome. This is basically how eukaryotes achieve to compact and facilitate the organization of the genome within the confines of the nucleus and temporally control the access to genetic elements. The nucleosome is composed of genomic DNA as well as two copies of each of the four canonical histones, H2A, H2B, H3 and H4. The access to genetic elements is regulated by various enzymatic activities including histone posttranslational modifications, chromatin remodeling and histone exchange by histone variants that alter the physical properties of the nucleosome or provide alternative sequences for posttranslational modifications and regulation.
Histone tails harbour multiple posttranslational modifications. Historically, it was proposed that these histone modifications could provide a code, termed appropriately the histone code,Citation1 which could dictate biological outcomes through protein-protein interactions with modification-specific binding proteins, broadly called readers. However, recent evidence suggests that a modification on the histone tail does not always lead to a pre-defined biological outcome, but depending on the context may even lead to opposite consequences. A notable paradigm involves the histone H3 trimethylated on lysine 4 (H3K4me3) mark, which is usually associated with transcriptional activity as it is present at the transcriptional start site (TSS) of most expressed genes.Citation2 In the context of DNA damage responses, H3K4me3 is read by the plant homeodomain (PHD) of the inhibitor of growth 2 (ING2) tumor suppressor, leading to transcriptional silencing of cell cycle genes.Citation3,Citation4 However, in response to genotoxic stress, H3K4me3 can also be read by ING4, which, through associated histone acetyltransferase activity, stimulates the transcription of cellular adhesion genes.Citation4,Citation5 Thus, the broader term chromatin signaling has been gaining popularity.Citation6-Citation8
Lysine methyltransferases (KMTs) are fundamental players in the regulation of chromatin signaling. This is emphasized by several reports showing that KMTs functional defects can lead to cancer,Citation9,Citation10 growth defects,Citation11 neurological disorders,Citation12 and other human pathologies. There are currently over 60 KMTs predicted in the latest human genome annotation. With the exception of DOT1LCitation13,Citation14 and the WRAD complex,Citation15 most KMTs harbor a predicted SET domain, which catalyzes the transfer of a methyl group from S-adenosylmethionine to the ε-amine on the side chain of lysine residue. Although predicted a few years ago,Citation16 ten members of the seven β-strand METhylTransferase-Like (METTL) family were recently characterized as KMTs.Citation17 Unlike other posttranslational modifications, lysine methylation occurs in three different flavors. Specifically, lysines can either be unmodified (K), mono (Kme1), di (Kme2) or trimethylated (Kme3). These incremental methylation states have the potential to lead to diverse biological outcomes through readers. These include Ankyrin, Chromo, MBT (Malignant Brain Tumor), PHD, PWWP (Proline-Tryptophan-Tryptophan-Proline), Tudor and WD40 domains.Citation18 The biological significance of aberrant chromatin signaling events is emphasized by the fact that several readers have clear links to cancer,Citation19-Citation21 suggesting a central role for lysine methylation in maintaining cellular homeostasis and in preventing neoplastic diseases.
Specific histone modifications appear to dictate whether or not a KMT can further modify its substrate. SET domain-containing methyltransferases seem to be particularly sensitive to the sequence and posttranslational modifications surrounding the target lysine site. I will explore within this short review the cross-talk between cis lysine methylation sites and other adjacent posttranslational modifications within histones H3 () and H4 as well as a few non-histone proteins.
Figure 1. Cross-talk on histone H3 N-terminus.The amino acid sequence of the histone tail of H3 is annotated to highlight the position of key modified residues. Black lines represents published cross-talk events. White lines represent putative cross-talk events. The dashed black line between H3R2 and H3K4 represents the antagonistic cross-talk between H3R2me2a and H3K4me3. The purple arrows between H3R2 and H3K4 highlight the permissive cross-talk between H3R2me2s and H3K4me3.
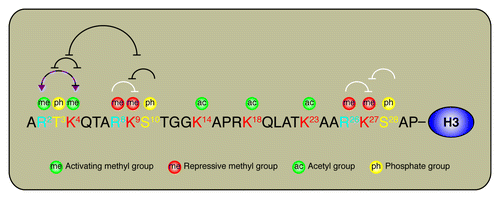
Regulation of H3 lysine 4 methylation (H3K4me)
The mixed lineage leukemia (MLL) protein lysine methyltransferase complex is evolutionarily conserved and mediates the methylation of H3K4. Chromosomal translocations of MLL are commonly found in leukemias and lead to aberrant expression of developmental and hematopoietic genes. Other KMTs that modify H3K4 include SETD1A and SET7.Citation22 The H3K4me1 modification marks enhancers,Citation23 while H3K4me3 surrounds transcriptional start sites and positively correlates with gene expression.Citation2
H3R2me2a prevents H3K4me
The asymmetric dimethylation of histone H3 arginine 2 (H3R2me2a) by the protein arginine methyltransferase PRMT6 precludes the methylation of H3K4 by the ASH2L/WDR5-containing MLL methyltransferase complex by preventing the WD40 repeat-containing WDR5 subunit from interacting with H3.Citation24 The H3R2me2a modification is conserved in Saccharomyces cerevisiae.Citation25 Interestingly, the H3R2me2a mark was shown to associate genome-wide with silenced chromatin and to prevent methylation of H3K4 by the Set1 lysine methyltransferase.Citation25
H3R2me2s facilitates H3K4me
The recently identified symmetrically dimethylated H3R2 (H3R2me2s) histone mark is not only found to overlap genome-wide with H3K4me3 in mouse, but it is conserved in Xenopus laevis, Drosophila melanogaster and Saccharomyces cerevisiae and detected in cis with H3K4me3 on the same histone tail.Citation26 Interestingly, the methylation of H3R2 requires both H3K4me3 as well as H3K4.Citation26 The arginine methyltransferases PRMT5 and PRMT7 were recently found to catalyze the formation of H3R2me2s.Citation27 Unlike the asymmetrically dimethylated form, H3R2me2s facilitates the interaction between H3 and the MLL complex subunit WDR5.Citation27 Thus, by enhancing MLL association with H3, WDR5 presumably facilitates H3K4me3 on H3R2me2s modified histones. Summarily, H3R2me2s facilitates H3K4me3 and conversely, H3K4me3 facilitates H3R2me2s.
H3T3ph prevents H3K4me3
The trimethylation of H3K4 prevents H3 phosphorylation on tyrosine 3 (H3T3ph) by haspin.Citation28 Interestingly, the opposite cross-talk effect was also be observed by H3T3ph on H3K4 methylation by MLL1.Citation29 Specifically, H3T3 is inserted in a defined structure within MLL1 SET domain.Citation29 The bulky and negatively charged phosphate group on H3T3ph would hypothetically lead to the repositioning of the threonine and likely change the orientation of the neighboring target lysine, thereby impairing MLL1 activity on H3K4.Citation29
H3K9me2/3 prevents H3K4me1
The SET7 lysine methyltransferase monomethylates the histone H3 on lysine 4 (H3K4me1), but also modifies non-histone proteins including the tumor suppressors p53Citation30 and pRB,Citation31 the hormone-responsive transcription factors estrogen receptor α (ERα)Citation32 and androgen receptor (AR),Citation33 the DNA methyltransferase DNMT1,Citation34 the histone deacetylase SIRT1,Citation35 as well as several other non-histone proteins.
Interestingly, SET7 has weaker activity on a H3K9me2 peptide relatively to the unmodified H3 peptide.Citation22 In addition, pre-methylation of H3 on K9 by the methyltransferase SUV39H1, which catalyzes the formation of H3K9me3, impaired SET7-dependent methylation on H3K4.Citation22 The crystal structure of H3-bound SET7 suggests that K9 from H3 is oriented toward the glutamic acid 271 (E271) of SET7. H3K9 and SET7E271 are presumably making electrostatic interactions.Citation36 Thus, H3K9me3 may affect these intermolecular interactions, preventing SET7 from methylating H3K4.
The mammalian homolog of the Drosophila melanogaster Trithorax group (TrxG) protein Ash1, ASH1L is a lysine methyltransferase that methylates histone H3 on possibly several sites, but is associated with active transcription.Citation37 Interestingly, the methylation activity of ASH1L on H3K4 is impaired by H3K9me3.Citation37 However, there is no structural evidence available to suggest a possible mechanism that could explain how H3K9me3 impairs ASH1L-mediated methylation of H3K4.
Biological consequences of H3K4me3 cross-talk
The presence of H3R2me2a in the body of genes and TSS prevents the deposition of H3K4me3 at silenced genes.Citation24 The absence of H3R2me2 and the presence of H3R2me2 at the TSS of actively transcribed genes facilitate the association of WDR5 with nucleosomes, thereby allowing trimethylation of H3K4.Citation27 Upon cell cycle arrest, the promoters of several transcriptional regulator genes are enriched with the H3R2me2 mark.Citation27 Although global levels of WDR5 are diminished in growth arrested cells, WDR5 is enriched at H3R2me2-marked promoters.Citation27
The H3R2me2s mark enhances the affinity of the RAG2 PHD domain for H3K4me3 by 20-fold.Citation26 Thus, the cross-talk between H3R2me2 and H3K4me3 possibly controls V(D)J recombination events mediated by RAG2 by enhancing the association of RAG2 at dually modified H3R2me2K4me3 chromatin loci, such as antigen receptor genes.
The MLL complex subunit Ash2L stimulates transcription that is driven by the TBP-associated factor TAF3 through methylation of H3K4 and thereby enhancing the interaction between H3K4me3 and the PHD domain of TAF3.Citation38 However, upon phosphorylation of H3T3 by haspin, Ash2L fails to stimulate TAF3-activated transcription.Citation39 In addition, H3T3 phosphorylation by haspin during mitosis is essential for proper alignment of metaphase chromosomes.Citation40 Hypothetically, phosphorylation of H3T3 by haspin during mitosis could prevent the deposition of H3K4me3 marks and the opening of condensed centromeric chromatin. Interestingly, pharmacological inhibition of haspin activity induces centrosome amplification, mitotic catastrophe and apoptosis.Citation41
Regulation of H3 lysine 9 methylation (H3K9me)
First identified as an H3K9-specific methyltransferase in 2002,Citation42 SETDB1 modifies H3K9Citation43 and ING2 in vitro.Citation44 Interestingly, SETDB1 catalytic activity is enhanced by an ATPase, mAM, which allows SETDB1 to convert H3K9me2 to H3K9me3.Citation45 There are several other H3K9-specific KMT, including SUV39H1,Citation46 SUV39H2Citation47 G9A,Citation48 and PRDM2.Citation49 Interestingly, G9A, GLP, SETDB1 and SUV39H1 form an enzymatic complex.Citation50 The H3K9me2 and H3K9me3 marks are enriched at the transcriptional start site of silenced genes, while H3K9me1 is found at transcribed promoters.Citation2
H3K4me3 prevents H3K9me3
Interestingly, the euchromatic mark H3K4me3 prevents methylation of H3K9 by SETDB1 as well as by the other H3K9-specific KMTs G9A and SUV39H1.Citation44 In vitro experimental approaches showed that H3K4me3 compromised methylation of H3K9 by SETDB1, G9A and SUV39H1.Citation44 Importantly, depletion of WDR82, an essential subunit of H3K4-specific KMT complexes,Citation51 led to severe reductions in H3K4me2/3 levels and concomitant increase in H3K9me3 levels in vivo,Citation44 arguing that methylation on the H3K4 site could inherently preclude H3K9 methylation, providing a passive mechanism for the segregation of the euchromatic and heterochromatic marks H3K4me3 and H3K9me3, respectively. It was independently reported that an un-specified methylation state of H3K4 impaired H3K9 methylation by SUV39H1 in vitro.Citation52
The structure of G9A reveals that histone H3 lysine 4 is buried in an acidic fold comprising the aspartic acids D1074 and D1088,Citation53 suggesting that the aspartic acid residues would confer electrostatic interactions with the positively charged H3K4 and that methylation of H3K4 could interfere with those interactions. Indeed, G9A activity on H3 is lower on H3K4me3, but the D1074A/D1088A G9A mutant has increased activity on H3K4me3 compared with the unmodified protein.Citation44 Hypothetically, the alanine mutations could provide additional space to accommodate the methyl groups of H3K4me3 into the acidic fold of G9A.
H3R8me potential effect on H3K9me3
The acetylation of H3K9 can prevent PRMT5 from methylating H3 arginine 8 (H3R8),Citation54 thus highlighting a potential cross-talk between H3R8me and H3K9me. Interestingly, the structure of G9A reveals that H3R8 is surrounded by three aspartic acids (D1074, D1078 and D1088) and that the amino groups on the side chain of H3R8 make electrostatic interactions with these three aspartic acid residues.Citation53 This acidic fold is shared by H3R8 and H3K9 where both H3 basic residues converge. The methylation of H3R8 by PRMT5 could undoubtedly sterically impede the proper insertion of H3 tail into the SET domain of G9A and prevent the methylation of H3K9.
H3S10ph prevents H3K9me3
Phosphorylation of H3 on serine 10 (H3S10ph) prevents methylation of H3K9 by G9ACitation55 and by SETDB1.Citation43 In addition, H3S10ph severely impairs methylation of H3K9 by SUV39H1 in vitro.Citation46 According to H3-bound G9A structure,Citation53 the OH group on the side chain of H3S10 makes electrostatic interactions with the arginine 1214 of G9A. Thus, phosphorylation of H3S10 could destabilize this interaction and possibly lead to poor KMT-substrate association and decreased H3K9 methylation. However, H3S10ph on already modified H3K9me3 does occur and is involved in regulating the association of the heterochromatin protein HP1 with H3K9me3.Citation56,Citation57 The impaired activity of G9A and SETDB1 on H3S10ph suggests that either H3S10 is phosphorylated only after the methylation of H3K9 or that another KMT is responsible for the catalysis of H3K9me3S10ph.
Biological consequences of H3K9me3 cross-talk
The H3S10ph mark prevents subsequent methylation of H3K9, but also prohibits the binding of the HP1 proteins to H3K9me3. Modulation of H3S10ph level by inhibiting or silencing the Aurora B kinase enhanced the association of HP1 proteins with mitotic chromosomes, suggesting a mechanism for the dissociation of HP1 proteins from chromatin during the M phase of the cell cycle.Citation56
Heterochromatin was proposed to be propagated via the association of HP1 proteins with H3K9me3, allowing further H3K9 methylation by the HP1-associated methyltransferase SUV39H1.Citation58 The cross-talk between H3K4me3 and H3K9me3 could provide a complementary mechanism to prevent the propagation of silenced chromatin states into transcriptionally active regions and vice versa.
Regulation of H3 lysine 27 methylation (H3K27me)
Although H3K27 surrounding amino acid sequence (ARKSA) is very similar to H3K9 (ARKST), only EZH1Citation59 and EZH2Citation60 were reported to catalyze the methylation of H3K27. The monomethyled H3K27me1 mark is enriched at actively transcribed promoters whereas the trimethylated H3K27me3 mark is associated with silenced promoters.Citation2
H3R26me and H3S28ph potential effect on H3K27me
Although the H3-bound EZH2 structure has not been solved yet, it is tempting to speculate based on the aforementioned cross-talk between H3K4 and H3K9 that either methylation of H3R26 by PRMT4Citation61 or phosphorylation of H3S28 by MSK1/2Citation62 could affect EZH2 association with H3 and its activity on H3K27. Mass spectrometric analysis detected the presence of H3S28ph on monomethylated and dimethylated H3K27, but not on trimethylated H3K27 peptides,Citation63 suggesting that H3K27me3 and H3S28ph are mutually exclusive modifications and that phosphorylation of H3S28 precludes the trimethylation of H3K27.
Regulation of H3 lysine 79 methylation (H3K79me)
H3T80ph potential effect on H3K79me
A few years ago the H3T80ph modification was detected by mass spectrometric analysis,Citation64 opening the possibility that, similarly to the cross-talk between H3S10ph and H3K9me3, H3T80ph could regulate the methylation of H3K79 by DOT1L. The crystal structure of the yeast ortholog of DOT1L, Dot1p, highlights an acidic cleft that could accommodate the basic charges surrounding H3K79, including R72 and R83.Citation65 Thus, the addition of a negatively charged phosphate group on H3S80 may affect the substrate-enzyme interaction. Indeed, the mutation of the acidic cleft of Dot1p abolished methyltransferase activity on H3K79.Citation65
Regulation of H4 lysine 20 methylation (H4K20me)
H4K16ac and H4K20me are antagonistic marks
Lysine methylation at H4K20 prevents the acetylation of H4K16 by the acetyltransferase p300 in vitro.Citation66 Interestingly, the inverse cross-talk was also observed whereby the H4K16ac mark prevents H4K20 methylation.Citation66 Although the H4K16ac and H4K20me marks are mutually exclusive, the H4K12ac mark was recently detected along with the novel H4K16me1 mark.Citation67 The structure of H4-bound SET8Citation68 reveals that H4K16 is surrounded by alanine 342 (A342), A346 and histidine 347 (H347), which are found at the carboxy terminal part of SET8 catalytic domain. The side chain of H347 makes hydrogen bonding with H4 peptide backbone.Citation68 In particular, the imidazole Nε2 atom of H347 and the backbone carbonyl of H4K16 are hydrogen bonding.Citation68 Thus, posttranslational modifications of H4K16 could alter these interactions and affect SET8 ability to methylate H4K20. Interestingly, an histidine to phenylalanine mutation at position 347 (H347F) led to increased substrate affinity.Citation68
Biological consequences of H4K20me2 cross-talk
Interestingly, the silencing of the histone acetyltransferase TIP60 decreased H4K16ac levels and induced the stabilization of 53BP1 association with H4K20me2 at DNA damage sites.Citation69 In addition, the inhibition of histone deacetylase activity by Trichostatin A led to enhanced H4K16ac levels and concomitant reduction in 53BP1 association with chromatin at DNA damage foci.Citation69 The interaction of 53BP1 tandem tudor domain (53BP1TT) with H4K20me2 is well-established.Citation70,Citation71 However, the acetylation of lysine 16 on H4K20me2 reduced the affinity of 53BP1TT for the mark. The acetylated form of H4 likely disrupts electrostatic interactions between H4K16 and an acidic patch in 53BP1 tandem tudor domain (amino acids E1549, D1550 and E1551).Citation69 The cross-talk between H4K16ac and H4K20me2 facilitates the dissociation of 53BP1 from chromatin at DNA damage breaks, allowing the recruitment of BRCA1 and homologous recombination repair.Citation69
H2A and the H2A variant H2AZ
The dual modification H2AK5ac K9me1 was recently detected by mass spectrometry.Citation67 Interestingly, H2AK5 aligns with H2AZK4 and H2AK9 aligns with H2AZK7. Both K4 and K7 on H2AZ were recently reported to be methylated by the methyltransferase SETD6.Citation72 However, acetylation and methylation of H2AZ are mutually exclusive modifications.Citation72 Given the similarities between the H2AK5K9 and H2AZK4K7 sequences, it seems likely that SETD6 could be responsible for the reported methylation of H2AK9.
Non-histone protein substrates cross-talk
Tumor suppressor p53
The p53 tumor suppressor protein is methylated on at least four lysine residues. SMYD2 mono-methylates p53 on lysine K370 (p53K370me1),Citation73,Citation74 while SET7 monomethylates p53 on lysine K372 (p53K372me1)Citation30 in the regulatory domain. The p53K372me1 modification leads to p53-dependent transcriptional activation.Citation30 Interestingly, the SET7-mediated p53K372me1 modification prevents methylation by SMYD2 on K370 and repression of p53 activities.Citation73 However, the SMYD2-mediated methylation of p53, p53K370me1, does not affect SET7 activity on p53.Citation73 The p53-bound SMYD2 structure was resolved and highlights several interactions between p53K372 and SMYD2 SET domain, including an hydrogen bond between p53K372 ε-amine group and the carbonyl of SMYD2 valine 215.Citation74 Therefore suggesting that p53K372me1 could sterically hinder the interaction of p53K372 with SMYD2 valine 215 and impair the methylation of K370 by SMYD2.Citation74
The tumor suppressor is also dimethylated by the G9A and G9A-like KMT GLP on lysine 373 (p53K373me2).Citation75 Interestingly, the aliphatic side chain of K373 from p53 is inserted within an aromatic cage of the SMYD2 catalytic domain that is lined by Y245, Y344, Y370 and Y374. The lysine K373 of p53 interacts directly with the side-chain of SMYD2Y344 through van der Waals interactions, whereas its ε-amine of p53K373 forms hydrogen bonds OH groups of Y370 and Y374.Citation74 Thus, G9A-mediated dimethylation of p53, p53K373me2, could hypothetically increase interactions with SMYD2 aromatic cage, as seen with the cation-π interactions-mediated increased affinity between ING4PHD and H3K4me3,Citation5 and lead to methylation of p53 at K370. This cooperative cross-talk between the two marks would be logical as both p53K370me1 and p53K373me2 inactivate p53 functions.Citation73,Citation75
Similarly to histones, lysine methylation of p53 not only involves cross-talk between the different modifications, but also serves as docking sites for readers. Specifically, the recognition of p53K370me2 by 53BP1 has the consequence of activating p53-dependent transcription.Citation76 The DNA damaging agent adriamycine activates a p53-dependent response and induces the methylation of p53K372 by SET7.Citation30 Chromatin signaling events leading to the methylation of p53 at K372 by SET7 could potentially prevent the modification of K370 by SMYD2 and diminish the association between p53 and 53BP1.
The p53 protein is also monomethylated at K382 (p53K382me1) by SET8.Citation77 In the context of DNA damage signaling induced by neocarzinostatin, the levels of p53K382me1 are reduced.Citation77 The methylation of p53 by SET8 leads to reduced p53-dependent expression of p21.Citation77 Interestingly, L3MBTL1 binds to p53K382me1 to silence the expression of p21 under normal conditions, but upon induction of DNA damage, p53 is relieved from L3MBTL1.Citation78 Summarily, the p53K382me1 mark provides a docking site for the transcriptional silencer L3MBTL1 and upon genotoxic stress, reduced p53K382me1 level are relieving L3MBTL1 from p53, thereby allowing p53-dependent transcriptional activation. Interestingly, the dimethylated (p53K382me2) form of p53 is induced by DNA damage and elicits the association with 53BP1.Citation79
DNA methyltransferase DNMT1
The DNA methyltransferase DNMT1 is lysine monomethylated on K142 by SET7.Citation34 Interestingly, the phosphorylation of DNMT1 at serine 143 by AKT1 interferes with the methylation of K142.Citation80 The DNMT1-bound SET7 structure reveals a polar interaction between DNMT1 S143 and K317 as well as van der Waals contact with L267 of SET7.Citation80 Therefore, S143ph should break the van der Waals contact with L267 and impair methylation of DNMT1K142.
Concluding remark
It is an exciting era for the field of chromatin signaling. With each new posttranslational modification being characterized, new doors for a potential cross-talk events and chromatin signaling networks open.
Acknowledgments
OB is supported by the Newcastle’s Biomedical Fellowship Programme, which is in part funded by the Wellcome Trust’s Institutional Strategic Support Fund.
Disclosure of Potential Conflict of Interests
No potential conflicts of interest were disclosed
References
- Jenuwein T, Allis CD. Translating the histone code. Science 2001; 293:1074 - 80; http://dx.doi.org/10.1126/science.1063127; PMID: 11498575
- Barski A, Cuddapah S, Cui K, Roh T-Y, Schones DE, Wang Z, et al. High-resolution profiling of histone methylations in the human genome. Cell 2007; 129:823 - 37; http://dx.doi.org/10.1016/j.cell.2007.05.009; PMID: 17512414
- Shi X, Hong T, Walter KL, Ewalt M, Michishita E, Hung T, et al. ING2 PHD domain links histone H3 lysine 4 methylation to active gene repression. Nature 2006; 442:96 - 9; PMID: 16728974
- Bua DJ, Binda O. The return of the INGs, histone mark sensors and phospholipid signaling effectors. Curr Drug Targets 2009; 10:418 - 31; http://dx.doi.org/10.2174/138945009788185112; PMID: 19442114
- Hung T, Binda O, Champagne KS, Kuo AJ, Johnson K, Chang HY, et al. ING4 mediates crosstalk between histone H3 K4 trimethylation and H3 acetylation to attenuate cellular transformation. Mol Cell 2009; 33:248 - 56; http://dx.doi.org/10.1016/j.molcel.2008.12.016; PMID: 19187765
- Sims RJ 3rd, Reinberg D. Is there a code embedded in proteins that is based on post-translational modifications?. Nat Rev Mol Cell Biol 2008; 9:815 - 20; http://dx.doi.org/10.1038/nrm2502; PMID: 18784729
- Lee J-S, Smith E, Shilatifard A. The language of histone crosstalk. Cell 2010; 142:682 - 5; http://dx.doi.org/10.1016/j.cell.2010.08.011; PMID: 20813257
- Henikoff S, Shilatifard A. Histone modification: cause or cog?. Trends Genet 2011; 27:389 - 96; http://dx.doi.org/10.1016/j.tig.2011.06.006; PMID: 21764166
- Albert M, Helin K. Histone methyltransferases in cancer. Semin Cell Dev Biol 2010; 21:209 - 20; http://dx.doi.org/10.1016/j.semcdb.2009.10.007; PMID: 19892027
- Schneider R, Bannister AJ, Kouzarides T. Unsafe SETs: histone lysine methyltransferases and cancer. Trends Biochem Sci 2002; 27:396 - 402; http://dx.doi.org/10.1016/S0968-0004(02)02141-2; PMID: 12151224
- Morishita M, di Luccio E. Cancers and the NSD family of histone lysine methyltransferases. Biochim Biophys Acta 2011; 1816:158 - 63; PMID: 21664949
- Ryu H, Lee J, Hagerty SW, Soh BY, McAlpin SE, Cormier KA, et al. ESET/SETDB1 gene expression and histone H3 (K9) trimethylation in Huntington’s disease. Proc Natl Acad Sci U S A 2006; 103:19176 - 81; http://dx.doi.org/10.1073/pnas.0606373103; PMID: 17142323
- Feng Q, Wang H, Ng HH, Erdjument-Bromage H, Tempst P, Struhl K, et al. Methylation of H3-lysine 79 is mediated by a new family of HMTases without a SET domain. Curr Biol 2002; 12:1052 - 8; http://dx.doi.org/10.1016/S0960-9822(02)00901-6; PMID: 12123582
- Min J, Feng Q, Li Z, Zhang Y, Xu R-M. Structure of the catalytic domain of human DOT1L, a non-SET domain nucleosomal histone methyltransferase. Cell 2003; 112:711 - 23; http://dx.doi.org/10.1016/S0092-8674(03)00114-4; PMID: 12628190
- Patel A, Vought VE, Dharmarajan V, Cosgrove MS. A novel non-SET domain multi-subunit methyltransferase required for sequential nucleosomal histone H3 methylation by the mixed lineage leukemia protein-1 (MLL1) core complex. J Biol Chem 2011; 286:3359 - 69; http://dx.doi.org/10.1074/jbc.M110.174524; PMID: 21106533
- Petrossian TC, Clarke SG. Uncovering the human methyltransferasome. Mol Cell Proteomics 2011; 10:M110.000976; http://dx.doi.org/10.1074/mcp.M110.000976; PMID: 20930037
- Cloutier P, Lavallée-Adam M, Faubert D, Blanchette M, Coulombe B. A newly uncovered group of distantly related lysine methyltransferases preferentially interact with molecular chaperones to regulate their activity. PLoS Genet 2013; 9:e1003210; http://dx.doi.org/10.1371/journal.pgen.1003210; PMID: 23349634
- Musselman CA, Lalonde M-È, Côté J, Kutateladze TG. Perceiving the epigenetic landscape through histone readers. Nat Struct Mol Biol 2012; 19:1218 - 27; http://dx.doi.org/10.1038/nsmb.2436; PMID: 23211769
- Baker LA, Allis CD, Wang GG. PHD fingers in human diseases: disorders arising from misinterpreting epigenetic marks. Mutat Res 2008; 647:3 - 12; http://dx.doi.org/10.1016/j.mrfmmm.2008.07.004; PMID: 18682256
- Musselman CA, Kutateladze TG. PHD fingers: epigenetic effectors and potential drug targets. Mol Interv 2009; 9:314 - 23; http://dx.doi.org/10.1124/mi.9.6.7; PMID: 20048137
- Bonasio R, Lecona E, Reinberg D. MBT domain proteins in development and disease. Semin Cell Dev Biol 2010; 21:221 - 30; http://dx.doi.org/10.1016/j.semcdb.2009.09.010; PMID: 19778625
- Wang H, Cao R, Xia L, Erdjument-Bromage H, Borchers C, Tempst P, et al. Purification and functional characterization of a histone H3-lysine 4-specific methyltransferase. Mol Cell 2001; 8:1207 - 17; http://dx.doi.org/10.1016/S1097-2765(01)00405-1; PMID: 11779497
- Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet 2007; 39:311 - 8; http://dx.doi.org/10.1038/ng1966; PMID: 17277777
- Guccione E, Bassi C, Casadio F, Martinato F, Cesaroni M, Schuchlautz H, et al. Methylation of histone H3R2 by PRMT6 and H3K4 by an MLL complex are mutually exclusive. Nature 2007; 449:933 - 7; http://dx.doi.org/10.1038/nature06166; PMID: 17898714
- Kirmizis A, Santos-Rosa H, Penkett CJ, Singer MA, Vermeulen M, Mann M, et al. Arginine methylation at histone H3R2 controls deposition of H3K4 trimethylation. Nature 2007; 449:928 - 32; http://dx.doi.org/10.1038/nature06160; PMID: 17898715
- Yuan C-C, Matthews AGW, Jin Y, Chen CF, Chapman BA, Ohsumi TK, et al. Histone H3R2 symmetric dimethylation and histone H3K4 trimethylation are tightly correlated in eukaryotic genomes. Cell reports 2012; 1:83 - 90; http://dx.doi.org/10.1016/j.celrep.2011.12.008; PMID: 22720264
- Migliori V, Müller J, Phalke S, Low D, Bezzi M, Mok WC, et al. Symmetric dimethylation of H3R2 is a newly identified histone mark that supports euchromatin maintenance. Nat Struct Mol Biol 2012; 19:136 - 44; http://dx.doi.org/10.1038/nsmb.2209; PMID: 22231400
- Eswaran J, Patnaik D, Filippakopoulos P, Wang F, Stein RL, Murray JW, et al. Structure and functional characterization of the atypical human kinase haspin. Proc Natl Acad Sci U S A 2009; 106:20198 - 203; http://dx.doi.org/10.1073/pnas.0901989106; PMID: 19918057
- Southall SM, Wong P-S, Odho Z, Roe SM, Wilson JR. Structural basis for the requirement of additional factors for MLL1 SET domain activity and recognition of epigenetic marks. Mol Cell 2009; 33:181 - 91; http://dx.doi.org/10.1016/j.molcel.2008.12.029; PMID: 19187761
- Chuikov S, Kurash JK, Wilson JR, Xiao B, Justin N, Ivanov GS, et al. Regulation of p53 activity through lysine methylation. Nature 2004; 432:353 - 60; http://dx.doi.org/10.1038/nature03117; PMID: 15525938
- Munro S, Khaire N, Inche A, Carr S, La Thangue NB. Lysine methylation regulates the pRb tumour suppressor protein. Oncogene 2010; 29:2357 - 67; http://dx.doi.org/10.1038/onc.2009.511; PMID: 20140018
- Subramanian K, Jia D, Kapoor-Vazirani P, Powell DR, Collins RE, Sharma D, et al. Regulation of estrogen receptor alpha by the SET7 lysine methyltransferase. Mol Cell 2008; 30:336 - 47; http://dx.doi.org/10.1016/j.molcel.2008.03.022; PMID: 18471979
- Gaughan L, Stockley J, Wang N, McCracken SR, Treumann A, Armstrong K, et al. Regulation of the androgen receptor by SET9-mediated methylation. Nucleic Acids Res 2010; 39;:4 1266 - 79; http://dx.doi.org/10.1093/nar/gkq861; PMID: 20959290
- Estève P-O, Chin HG, Benner J, Feehery GR, Samaranayake M, Horwitz GA, et al. Regulation of DNMT1 stability through SET7-mediated lysine methylation in mammalian cells. Proc Natl Acad Sci U S A 2009; 106:5076 - 81; http://dx.doi.org/10.1073/pnas.0810362106; PMID: 19282482
- Liu X, Wang D, Zhao Y, Tu B, Zheng Z, Wang L, et al. Methyltransferase Set7/9 regulates p53 activity by interacting with Sirtuin 1 (SIRT1). Proc Natl Acad Sci U S A 2011; 108:1925 - 30; http://dx.doi.org/10.1073/pnas.1019619108; PMID: 21245319
- Xiao B, Jing C, Wilson JR, Walker PA, Vasisht N, Kelly G, et al. Structure and catalytic mechanism of the human histone methyltransferase SET7/9. Nature 2003; 421:652 - 6; http://dx.doi.org/10.1038/nature01378; PMID: 12540855
- Gregory GD, Vakoc CR, Rozovskaia T, Zheng X, Patel S, Nakamura T, et al. Mammalian ASH1L is a histone methyltransferase that occupies the transcribed region of active genes. Mol Cell Biol 2007; 27:8466 - 79; http://dx.doi.org/10.1128/MCB.00993-07; PMID: 17923682
- Vermeulen M, Mulder KW, Denissov S, Pijnappel WWMP, van Schaik FMA, Varier RA, et al. Selective anchoring of TFIID to nucleosomes by trimethylation of histone H3 lysine 4. Cell 2007; 131:58 - 69; http://dx.doi.org/10.1016/j.cell.2007.08.016; PMID: 17884155
- Varier RA, Outchkourov NS, de Graaf P, van Schaik FMA, Ensing HJL, Wang F, et al. A phospho/methyl switch at histone H3 regulates TFIID association with mitotic chromosomes. EMBO J 2010; 29:3967 - 78; http://dx.doi.org/10.1038/emboj.2010.261; PMID: 20953165
- Dai J, Sultan S, Taylor SS, Higgins JMG. The kinase haspin is required for mitotic histone H3 Thr 3 phosphorylation and normal metaphase chromosome alignment. Genes Dev 2005; 19:472 - 88; http://dx.doi.org/10.1101/gad.1267105; PMID: 15681610
- Huertas D, Soler M, Moreto J, Villanueva A, Martinez A, Vidal A, et al. Antitumor activity of a small-molecule inhibitor of the histone kinase Haspin. Oncogene 2012; 31:1408 - 18; http://dx.doi.org/10.1038/onc.2011.335; PMID: 21804608
- Yang L, Xia L, Wu DY, Wang H, Chansky HA, Schubach WH, et al. Molecular cloning of ESET, a novel histone H3-specific methyltransferase that interacts with ERG transcription factor. Oncogene 2002; 21:148 - 52; http://dx.doi.org/10.1038/sj.onc.1204998; PMID: 11791185
- Schultz DC, Ayyanathan K, Negorev D, Maul GG, Rauscher FJ 3rd. SETDB1: a novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes Dev 2002; 16:919 - 32; http://dx.doi.org/10.1101/gad.973302; PMID: 11959841
- Binda O, LeRoy G, Bua DJ, Garcia BA, Gozani O, Richard S. Trimethylation of histone H3 lysine 4 impairs methylation of histone H3 lysine 9: regulation of lysine methyltransferases by physical interaction with their substrates. Epigenetics 2010; 5:767 - 75; http://dx.doi.org/10.4161/epi.5.8.13278; PMID: 21124070
- Wang H, An W, Cao R, Xia L, Erdjument-Bromage H, Chatton B, et al. mAM facilitates conversion by ESET of dimethyl to trimethyl lysine 9 of histone H3 to cause transcriptional repression. Mol Cell 2003; 12:475 - 87; http://dx.doi.org/10.1016/j.molcel.2003.08.007; PMID: 14536086
- Rea S, Eisenhaber F, O’Carroll D, Strahl BD, Sun Z-W, Schmid M, et al. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature 2000; 406:593 - 9; http://dx.doi.org/10.1038/35020506; PMID: 10949293
- O’Carroll D, Scherthan H, Peters AH, Opravil S, Haynes AR, Laible G, et al. Isolation and characterization of Suv39h2, a second histone H3 methyltransferase gene that displays testis-specific expression. Mol Cell Biol 2000; 20:9423 - 33; http://dx.doi.org/10.1128/MCB.20.24.9423-9433.2000; PMID: 11094092
- Tachibana M, Sugimoto K, Fukushima T, Shinkai Y. Set domain-containing protein, G9a, is a novel lysine-preferring mammalian histone methyltransferase with hyperactivity and specific selectivity to lysines 9 and 27 of histone H3. J Biol Chem 2001; 276:25309 - 17; http://dx.doi.org/10.1074/jbc.M101914200; PMID: 11316813
- Kim K-C, Geng L, Huang S. Inactivation of a histone methyltransferase by mutations in human cancers. Cancer Res 2003; 63:7619 - 23; PMID: 14633678
- Fritsch L, Robin P, Mathieu JRR, Souidi M, Hinaux H, Rougeulle C, et al. A subset of the histone H3 lysine 9 methyltransferases Suv39h1, G9a, GLP, and SETDB1 participate in a multimeric complex. Mol Cell 2010; 37:46 - 56; http://dx.doi.org/10.1016/j.molcel.2009.12.017; PMID: 20129054
- Wu M, Wang PF, Lee JS, Martin-Brown S, Florens L, Washburn M, et al. Molecular regulation of H3K4 trimethylation by Wdr82, a component of human Set1/COMPASS. Mol Cell Biol 2008; 28:7337 - 44; http://dx.doi.org/10.1128/MCB.00976-08; PMID: 18838538
- Nishioka K, Chuikov S, Sarma K, Erdjument-Bromage H, Allis CD, Tempst P, et al. Set9, a novel histone H3 methyltransferase that facilitates transcription by precluding histone tail modifications required for heterochromatin formation. Genes Dev 2002; 16:479 - 89; http://dx.doi.org/10.1101/gad.967202; PMID: 11850410
- Wu H, Min J, Lunin VV, Antoshenko T, Dombrovski L, Zeng H, et al. Structural biology of human H3K9 methyltransferases. PLoS One 2010; 5:e8570; http://dx.doi.org/10.1371/journal.pone.0008570; PMID: 20084102
- Pal S, Vishwanath SN, Erdjument-Bromage H, Tempst P, Sif S. Human SWI/SNF-associated PRMT5 methylates histone H3 arginine 8 and negatively regulates expression of ST7 and NM23 tumor suppressor genes. Mol Cell Biol 2004; 24:9630 - 45; http://dx.doi.org/10.1128/MCB.24.21.9630-9645.2004; PMID: 15485929
- Duan Q, Chen H, Costa M, Dai W. Phosphorylation of H3S10 blocks the access of H3K9 by specific antibodies and histone methyltransferase. Implication in regulating chromatin dynamics and epigenetic inheritance during mitosis. J Biol Chem 2008; 283:33585 - 90; http://dx.doi.org/10.1074/jbc.M803312200; PMID: 18835819
- Fischle W, Tseng BS, Dormann HL, Ueberheide BM, Garcia BA, Shabanowitz J, et al. Regulation of HP1-chromatin binding by histone H3 methylation and phosphorylation. Nature 2005; 438:1116 - 22; http://dx.doi.org/10.1038/nature04219; PMID: 16222246
- Hirota T, Lipp JJ, Toh B-H, Peters J-M. Histone H3 serine 10 phosphorylation by Aurora B causes HP1 dissociation from heterochromatin. Nature 2005; 438:1176 - 80; http://dx.doi.org/10.1038/nature04254; PMID: 16222244
- Lachner M, O’Carroll D, Rea S, Mechtler K, Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 2001; 410:116 - 20; http://dx.doi.org/10.1038/35065132; PMID: 11242053
- Shen X, Liu Y, Hsu Y-J, Fujiwara Y, Kim J, Mao X, et al. EZH1 mediates methylation on histone H3 lysine 27 and complements EZH2 in maintaining stem cell identity and executing pluripotency. Mol Cell 2008; 32:491 - 502; http://dx.doi.org/10.1016/j.molcel.2008.10.016; PMID: 19026780
- Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, et al. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science 2002; 298:1039 - 43; http://dx.doi.org/10.1126/science.1076997; PMID: 12351676
- Schurter BT, Koh SS, Chen D, Bunick GJ, Harp JM, Hanson BL, et al. Methylation of histone H3 by coactivator-associated arginine methyltransferase 1. Biochemistry 2001; 40:5747 - 56; http://dx.doi.org/10.1021/bi002631b; PMID: 11341840
- Zhong S, Jansen C, She QB, Goto H, Inagaki M, Bode AM, et al. Ultraviolet B-induced phosphorylation of histone H3 at serine 28 is mediated by MSK1. J Biol Chem 2001; 276:33213 - 9; http://dx.doi.org/10.1074/jbc.M103973200; PMID: 11441012
- Bonenfant D, Towbin H, Coulot M, Schindler P, Mueller DR, van Oostrum J. Analysis of dynamic changes in post-translational modifications of human histones during cell cycle by mass spectrometry. Mol Cell Proteomics 2007; 6:1917 - 32; http://dx.doi.org/10.1074/mcp.M700070-MCP200; PMID: 17644761
- Vermeulen M, Eberl HC, Matarese F, Marks H, Denissov S, Butter F, et al. Quantitative interaction proteomics and genome-wide profiling of epigenetic histone marks and their readers. Cell 2010; 142:967 - 80; http://dx.doi.org/10.1016/j.cell.2010.08.020; PMID: 20850016
- Sawada K, Yang Z, Horton JR, Collins RE, Zhang X, Cheng X. Structure of the conserved core of the yeast Dot1p, a nucleosomal histone H3 lysine 79 methyltransferase. J Biol Chem 2004; 279:43296 - 306; http://dx.doi.org/10.1074/jbc.M405902200; PMID: 15292170
- Nishioka K, Rice JC, Sarma K, Erdjument-Bromage H, Werner J, Wang Y, et al. PR-Set7 is a nucleosome-specific methyltransferase that modifies lysine 20 of histone H4 and is associated with silent chromatin. Mol Cell 2002; 9:1201 - 13; http://dx.doi.org/10.1016/S1097-2765(02)00548-8; PMID: 12086618
- Tan M, Luo H, Lee S, Jin F, Yang JS, Montellier E, et al. Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell 2011; 146:1016 - 28; http://dx.doi.org/10.1016/j.cell.2011.08.008; PMID: 21925322
- Couture J-F, Collazo E, Brunzelle JS, Trievel RC. Structural and functional analysis of SET8, a histone H4 Lys-20 methyltransferase. Genes Dev 2005; 19:1455 - 65; http://dx.doi.org/10.1101/gad.1318405; PMID: 15933070
- Tang J, Cho NW, Cui G, Manion EM, Shanbhag NM, Botuyan MV, et al. Acetylation limits 53BP1 association with damaged chromatin to promote homologous recombination. Nat Struct Mol Biol 2013; 20:317 - 25; http://dx.doi.org/10.1038/nsmb.2499; PMID: 23377543
- Botuyan MV, Lee J, Ward IM, Kim J-E, Thompson JR, Chen J, et al. Structural basis for the methylation state-specific recognition of histone H4-K20 by 53BP1 and Crb2 in DNA repair. Cell 2006; 127:1361 - 73; http://dx.doi.org/10.1016/j.cell.2006.10.043; PMID: 17190600
- Sanders SL, Portoso M, Mata J, Bähler J, Allshire RC, Kouzarides T. Methylation of histone H4 lysine 20 controls recruitment of Crb2 to sites of DNA damage. Cell 2004; 119:603 - 14; http://dx.doi.org/10.1016/j.cell.2004.11.009; PMID: 15550243
- Binda O, Sevilla A, Leroy G, Lemischka IR, Garcia BA, Richard S. SETD6 monomethylates H2AZ on lysine 7 and is required for the maintenance of embryonic stem cell self-renewal. Epigenetics 2013; 8:177 - 83; http://dx.doi.org/10.4161/epi.23416; PMID: 23324626
- Huang J, Perez-Burgos L, Placek BJ, Sengupta R, Richter M, Dorsey JA, et al. Repression of p53 activity by Smyd2-mediated methylation. Nature 2006; 444:629 - 32; http://dx.doi.org/10.1038/nature05287; PMID: 17108971
- Wang L, Li L, Zhang H, Luo X, Dai J, Zhou S, et al. Structure of human SMYD2 protein reveals the basis of p53 tumor suppressor methylation. J Biol Chem 2011; 286:38725 - 37; http://dx.doi.org/10.1074/jbc.M111.262410; PMID: 21880715
- Huang J, Dorsey J, Chuikov S, Pérez-Burgos L, Zhang X, Jenuwein T, et al. G9a and Glp methylate lysine 373 in the tumor suppressor p53. J Biol Chem 2010; 285:9636 - 41; http://dx.doi.org/10.1074/jbc.M109.062588; PMID: 20118233
- Huang J, Sengupta R, Espejo AB, Lee MG, Dorsey JA, Richter M, et al. p53 is regulated by the lysine demethylase LSD1. Nature 2007; 449:105 - 8; http://dx.doi.org/10.1038/nature06092; PMID: 17805299
- Shi X, Kachirskaia I, Yamaguchi H, West LE, Wen H, Wang EW, et al. Modulation of p53 function by SET8-mediated methylation at lysine 382. Mol Cell 2007; 27:636 - 46; http://dx.doi.org/10.1016/j.molcel.2007.07.012; PMID: 17707234
- West LE, Roy S, Lachmi-Weiner K, Hayashi R, Shi X, Appella E, et al. The MBT repeats of L3MBTL1 link SET8-mediated p53 methylation at lysine 382 to target gene repression. J Biol Chem 2010; 285:37725 - 32; http://dx.doi.org/10.1074/jbc.M110.139527; PMID: 20870725
- Kachirskaia I, Shi X, Yamaguchi H, Tanoue K, Wen H, Wang EW, et al. Role for 53BP1 Tudor domain recognition of p53 dimethylated at lysine 382 in DNA damage signaling. J Biol Chem 2008; 283:34660 - 6; http://dx.doi.org/10.1074/jbc.M806020200; PMID: 18840612
- Estève P-O, Chang Y, Samaranayake M, Upadhyay AK, Horton JR, Feehery GR, et al. A methylation and phosphorylation switch between an adjacent lysine and serine determines human DNMT1 stability. Nat Struct Mol Biol 2011; 18:42 - 8; http://dx.doi.org/10.1038/nsmb.1939; PMID: 21151116