Abstract
DNA methylation has been studied in many eukaryotic organisms, in particular vertebrates, and was implicated in developmental and phenotypic variations. Little is known about the role of DNA methylation in invertebrates, although insects are considered as excellent models for studying the evolution of DNA methylation. In the red flour beetle, Tribolium castaneum (Tenebrionidae, Coleoptera), no evidence of DNA methylation has been found till now. In this paper, a cytosine methylation in Tribolium castaneum embryos was detected by methylation sensitive restriction endonucleases and immuno-dot blot assay. DNA methylation in embryos is followed by a global demethylation in larvae, pupae and adults. DNA demethylation seems to proceed actively through 5-hydroxymethylcytosine, most probably by the action of TET enzyme. Bisulfite sequencing of a highly abundant satellite DNA located in pericentromeric heterochromatin revealed similar profile of cytosine methylation in adults and embryos. Cytosine methylation was not only restricted to CpG sites but was found at CpA, CpT and CpC sites. In addition, complete cytosine demethylation of heterochromatic satellite DNA was induced by heat stress. The results reveal existence of DNA methylation cycling in T. castaneum ranging from strong overall cytosine methylation in embryos to a weak DNA methylation in other developmental stages. Nevertheless, DNA methylation is preserved within heterochromatin during development, indicating its role in heterochromatin formation and maintenance. It is, however, strongly affected by heat stress, suggesting a role for DNA methylation in heterochromatin structure modulation during heat stress response.
Introduction
Methylation of cytosine is widely spread in eukaryotes and seems to be important for transcriptional regulation, heterochromatin formation and transposon inactivation.Citation1 In mammals, cytosine methylation represents the epigenetic mark responsible for genomic imprinting.Citation2 DNA methylation occurs in vertebrates predominantly at CpG sites and approximately 60% to 90% of CpG dinucleotides are methylated globally throughout the genome.Citation3 Presence of 5-methylcytosine has been reported in several insects (for a review see reference Citation4), but the number of methylated CpGs is significantly lower than in vertebrates, as revealed by the whole genome methylation analysis of the silk worm Bombyx mori,Citation5 and honey bee Apis mellifera.Citation6 In the honey bee A. mellifera, DNA methylation has been implicated in the regulation of phenotypic plasticity and seems to determine the developmental fates in response to environmental signals.Citation7 However, further research is required to fully understand the role of DNA methylation in this evolutionarily diverse class.Citation8,Citation9
The beetle Tribolium castaneum is an important pest of stored products and a powerful model organism for the study of insect development. Until now, no evidence of genomic methylation in T. castaneum adults was demonstrated.Citation10 In addition to T. castaneum, a near-total lack of DNA methylation was reported in the fruit fly Drosophila melanogaster as well as in mosquito Anopheles gambiae.Citation11 The currently available genome sequence of T. castaneum seems to encode for DNA methyltransferases DNMT1 and DNMT2 and for MBD (methyl-CpG-binding domain) proteins.Citation12 DNMT1 and DNMT3 are generally considered necessary to a functional DNA methylation system and the absence of DNMT3 in T. castaneum is proposed to be associated with the loss of DNA methylation.Citation13 Intriguingly, the same set of DNMTs as in T. castaneum is found in the silk worm B. mori, but this species exhibits genomic methylation, although at low level.Citation5
In insects, DNA methylation seems to be specific for intragenic regions while transposons and other intergenic repetitive elements are largely unmethylated.Citation3,Citation10,Citation14 However, there are examples of strongly methylated interspersed repetitive DNA and genes in the stick insect Medauroidea extradentata.Citation15 The genome of the Lepidoptera Mamestra brassicae has a vertebrate-like content of methylcytosine but, using the methylation sensitive restriction enzyme assay, no evidence of CpG methylation was demonstrated on repetitive DNA such as transposons, rDNA and the MBSAT1 satellite DNA.Citation16,Citation17 In the aphid Aphis nerii (Hemiptera), heterochromatin seems to be assembled and condensed without involvement of DNA methylation, which is restricted to euchromatin.Citation18 These results seem to argue against a classical role for DNA methylation in transposon silencing and in control of heterochromatin formation via satellite DNA methylation. Furthermore, the intragenic DNA methylation characteristic of insects might be involved in the regulation of gene expression through mRNA splicing.Citation19
In plants and mammals, DNA demethylation takes place in germ cells and early embryos during epigenetic reprogramming, representing a key mechanism during early development.Citation14 Demethylation of mammalian promoters is also prone to dynamic changes influencing gene expression.Citation20,Citation21 In insects, the role of DNA demethylation is still unexplored. It is unknown whether DNA demethylation is critical during development and if methylation cycling exists in insects. Here we demonstrate a global cytosine methylation in the red flour beetle T. castaneum in the embryonic stage followed by demethylation during the first step of metamorphosis and in the remaining developmental phases. In order to study if DNA methylation plays a role in heterochromatin formation and regulation in T. castaneum, we performed DNA methylation analysis within highly abundant pericentromeric satellite DNA TCAST, which makes 35% of the genome,Citation22 using bisulfite sequencing. Constant methylation of cytosine within heterochromatic satellite DNA was detected in all developmental stages. In heat-shock treated beetles, overexpression of satellite DNA was accompanied by strong satellite DNA demethylation.
Our results show, for the first time, the presence of DNA methylation in T. castaneum embryos. Using sensitive bisulfite sequencing, we revealed cytosine methylation within heterochromatic DNA in embryos and adults. This methylation is modulated by environmental stress.
Results
Detection of DNA methylation in T. castaneum genome
Using methylation sensitive restriction endonucleases (MSRE), a comparative differential cleavage pattern between genomic DNAs from different developmental stages of T. castaneum was detected. Digestion of genomic DNAs using the methylation sensitive restriction enzyme Hin6I (GCGC) revealed an almost complete digestion pattern in larvae, pupae and adults relative to embryos (). The comparison of digestion profiles indicates significant level of cytosine methylation at CpG sites in embryos relative to the low methylation level in other three developmental phases.
Figure 1. (A) Agarose gel electrophoresis of equivalent amounts of genomic DNA from 3 d old embryos, larvae, pupae and adults digested using: Hin6I restriction enzyme (lanes: 1-embryos with plasmid pUC18R as a control of digestion, 2-embryos, 3-larvae, 4-pupae, 5 -adults). (B) Methylation-sensitive restriction enzyme HpaII (lanes: 2-embryos, 5-adults) and with the methylation-insensitive restriction enzyme MspI (lanes: 3-embryos, 6- adults). Undigested DNA from embryos and adults is present in lanes 1 and 4, respectively. (C) Methylation specific GlaI enzyme digestion of DNAs from embryos (lane 2) and from adults (lane 4) compared with the undigested controls (lanes: 1-embryos, 3-adults). (D)PvuRts1I enzyme digestion of DNA from embryos, larvae, pupae and adults (lanes 1, 2, 3, 4 respectively) compared with the undigested controls (ND). The tracks labeled L contain GeneRuler Ladder mix (Fermentas).
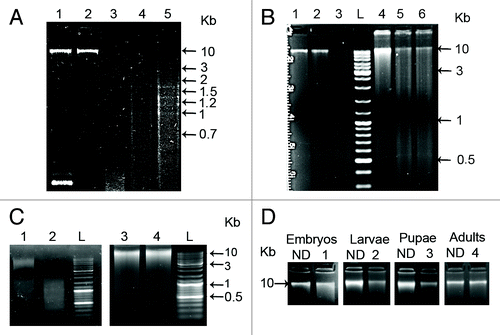
The same result was detected using a pair of isoschizomers, HpaII (sensitive to methylation) and MspI (methylation insensitive), that recognize the target sequence CCGG. Genomic DNA from embryos was significantly digested by MspI (, lane 3) relative to HpaII (, lane 2), which showed profile very similar to the undigested control (, lane 1). Unlike embryos, no obvious differences were found in adults between the digestion patterns of HpaII and MspI (, lanes 5 and 6), and both enzymes revealed significant digestion relative to the control (, lane 4). These results argue again in favor of a significantly higher CpG methylation in embryos than in larvae, pupae and adults.
In addition, to confirm the developmental stage-specific CpG methylation, genomic DNAs from embryos and adults were digested with the methylation-specific enzyme GlaI, an isoschizomer of Hin6I, which cleaves only C5-methylated DNA and does not cut unmodified DNA and DNA with N4-methylcytosines (). The results show significant digestion of embryonic DNA by GlaI (, lane 2) relative to the undigested control DNA (, lane 1), contrary to adult DNA, which is poorly digested by GlaI relative to the control (, lanes 3 and 4). As a positive control of complete digestion with GlaI, a methylated linearized plasmid pHspAI2/GsaI of 4,118 bp in size, containing one GlaI site, was used (not shown). These results confirm an overall CpG methylation in GCGC sites restricted to embryonic stage and a CpG demethylation in adult stage. Identical results were obtained using embryos of different age (3–24 h) and adults of different gender (not shown), indicating an early occurrence of CpG methylation in zygote and excluding overall gender-specific CpG methylation.
Cytosine demethylation observed during T. castaneum development can occur passively in the absence of methylation of newly synthesized DNA strands during replication or actively, via direct removal of a methyl group. The TET family of enzymes are considered to be responsible for active demethylation process, by catalyzing the production of 5- hydroxymethylcytosine (5- hmC) from 5- methylcytosine (5-mC).Citation23 A hypothetical protein, TcasGA2_TC013798, present in Tribolium castaneum GenBank under accession number EFA03694.1, shows conserved domains highly homologous to TET enzymes of different organisms. The presence of 5-hmC in T. castaneum genomic DNA was followed using PvuRts1I restriction enzyme (recognition sequence: hmCN11–12/N9–10G), which cleaves 5-hmC DNA specifically and does not digest 5-methylcytosine residues or unmethylated DNA. Genomic DNA from all developmental stages was digested with PvuRts1I enzyme and the digested samples were compared with the same amount of undigested DNA (). Strong evidence of digestion was detected only in larvae and pupae DNA while samples corresponding to embryos and adults DNA were either not digested or very poorly digested (). This experiment using PvuRts1I restriction enzyme indicates, for the first time, the presence of 5-hydroxymethylcytosine in T. castaneum, with strong indication of its presence limited mainly to larvae and pupae developmental stages.
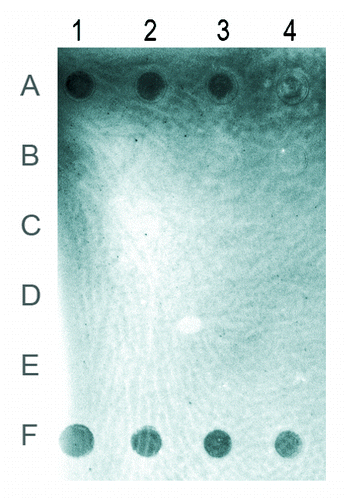
The presence of cytosine methylation in T. castaneum genomic DNA was also checked by immunological reaction, using an antibody specific for 5-methylcytosine. The immuno-dot blot assay performed using serial dilutions of genomic DNA, from 100 ng to 12.5 ng, from all stages of development, revealed strong signal intensity in embryos (). In larvae, weak signal is visible only at a dot corresponding to the highest DNA amount (, dot 1B), indicating that the level of 5-mC in this developmental phase is at least ten times lower than the level detected in embryos. In pupae and adults, no signal was detectable, which could be due to a very low amount of 5-mC that is below the sensitivity of the immuno-dot blot assay.
Figure 2. Immuno-dot blot using antibody specific for 5-methylcytosine. Rows (A) to (D) represent genomic DNAs from embryos, larvae, pupae and adults, respectively. Row (E), unmethylated DNA used as a negative control. Row (F), 5-methylcytosine DNA used as a positive control. The amount of genomic DNA in the dots of columns 1–4 was 100, 50, 25 and 12.5 ng. The brightness of the image was modified using the Curve function in Photoshop.
DNA methylation within heterochromatic satellite DNA
MSRE (methylation sensitive restriction enzyme) analysis
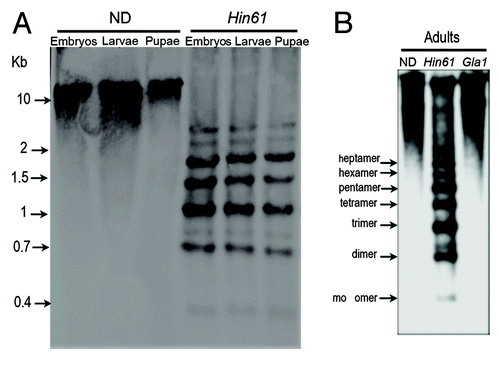
Large blocks of pericentromeric heterochromatin are present on all 20 chromosomes of T. castaneum and are composed of TCAST satellite, which makes up 35% of the whole genome.Citation24 Although genome-wide analysis of DNA methylation in T. castaneum has been performed,Citation10 the analysis excluded satellite DNA due to the inability to map tandem repeats to chromosomes. Using the methylation sensitive restriction enzyme Hin6I combined with Southern hybridization, we specifically checked the level of CpG methylation within highly abundant TCAST satellite in all developmental stages. TCAST satellite DNA is composed of two highly homologous subfamilies, Tcast1a and Tcast1b, that are principally organized into composite arrays of Tcast1a+Tcast1b.Citation22 Because a part of Tcast1a repeats include a recognition site for Hin6I, digestion of T. castaneum genomic DNA with Hin6I enzyme produces a specific hybridization pattern using a TCAST satellite DNA as a probe (). Comparison of the restriction pattern of Hin6I did not reveal any significant difference among DNA from embryos, larvae, pupae and adults (). The presence of oligomers (trimers, tetramers, etc.) also indicates that many Tcast1a repeats have the Hin6I sites inactivated by mutation and/or methylation. The similarity among patterns indicates that, despite significantly higher overall CpG methylation in embryos, methylation at GCGC sites within heterochromatic Tcast1a satellite DNA is similar in all developmental stages, including embryos. Different from the Hin6I hybridization pattern, digestion of adult DNA with GlaI, followed by hybridization with a TCAST probe, gave only high molecular weight signal (), indicating low methylation at GCGC sites within Tcast1a repeats of TCAST satellite DNA.
Figure 3. Southern hybridization analysis of T. castaneum genomic DNA using as a hybridization probe mixture of biotin labeled Tcast1a and Tcast1b monomers. (A) Equivalent amounts of genomic DNA from embryos (three days old), larvae and pupae were digested using Hin6I and compared with undigested DNA. (B) Equivalent amounts of genomic DNA from adults were digested with Hin6I and GlaI and compared with undigested DNA.
Bisulfite sequencing
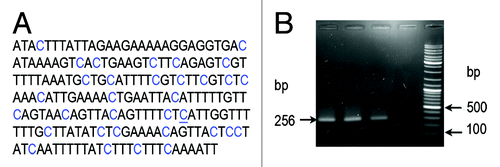
Cytosine methylation within heterochromatic TCAST satellite DNA was further investigated using a more sensitive method of bisulfite sequencing. The classical bisulfite sequencing method consists of three steps: bisulfite chemical treatment that converts all not-methylated cytosines into uracils but does not modify 5-mC and 5-hmC; PCR amplification, where all uracils are converted into thymines; and a last step in which PCR products purified from the gel are cloned into an appropriate vector and used for sequencing analysis. The primers used for TCAST methylation analysis were designed by MethPrimer software using Tcast1a+Tcast1b heterodimer sequence. TCAST methprimers are expected to amplify a Tcast1b subregion of 265 base pairs in size, containing 33 cytosines (). The primers are able to anneal selectively to target regions containing exclusively not-methylated cytosine. As a control, the TCAST methylprimer pair was tested for PCR amplification using genomic DNA not modified by bisulfite. As expected, there was no amplification (), indicating that TCAST methylprimers can function only on bisulfite modified genomic DNA.
Figure 4.(A) A region of TCAST satellite sequence of 265 bp used for cytosine methylation analysis by bisulfite treatment. Positions of 33 cytosines are marked in blue, and a single position where no methylation was detected is underlined. (B) PCR amplification using TCAST methylprimers and bisulfite treated genomic DNAs (lanes 1, 2, 3) and untreated, control DNA (lane 4). In lanes 1–3 genomic DNA was isolated from T. castaneum adults not subjected to heat shock and heat shocked for 3 h and 20 h, respectively. The size of amplicon is 265 bp. The lane 5 contains GeneRuler Ladder mix (Fermentas).
The level of cytosine methylation within heterochromatic TCAST satellite DNA was tested on embryo and adult DNA. Since it was previously observed that the level of expression of TCAST satellite DNA is significantly increased after heat shock treatment,Citation25 we also checked if the level of methylation of TCAST satellite was affected by heat shock. Using TCAST-methprimers, we amplified bisulfite treated DNA from embryos and adults not subjected to heat shock, as well as from adults heat shocked for 3 h or 20 h (). Bands were isolated from the gel and used for direct sequencing to investigate the overall methylation status of the repeats in the genome. This procedure was previously successfully applied to determine the average genomic sequence of different satellite DNA, including TCAST satellite DNA,Citation22,Citation26,Citation27 and was, for the first time, tested using bisulfite converted genomic DNA. Each amplification product originates from a large number of repeats and, when a methylcytosine is found at a specific position and cytosine at others, the electropherogram will show peaks corresponding to a mixture of cytosine and thymine. If cytosines are homogeneously methylated or unmethylated at some positions within the repeats, peaks of either cytosine or thymine will be visible. The electropherograms of the region of analyzed TCAST sequence are shown on . The analysis of adult DNA revealed partial presence of 5-mC and/or 5-hmC at 32 out of 33 total cytosine positions, while at a single CpT position no methylation was detected (). Almost the same methylation profile was obtained for embryo DNA (not shown). Methylation is not restricted to CpG but is also found at CpA, CpT and CpC sites. The results also indicate a progressive cytosine demethylation after heat shock treatment (). After 3 h of heat stress at 40°C, a significant decrease in cytosine methylation was observed at all positions relative to the control, while after 20 h of heat stress all cytosines were almost completely demethylated ()
Figure 5. Electropherograms of a part of TCAST satellite sequence after bisulfite treatment of genomic DNAs isolated from adults (A), from adults heat shocked for 3 h (B) and 20 h (C). Red and blue peaks indicate thymine and cytosine, respectively. Each cytosine position is indicated by yellow column and a single position within TCAST region where the cytosine is present in completely unmethylated state is marked and underlined. For each sample, the methylation status of polyubiquitin and cinnabar gene promoters were also checked and the results were identical for all samples, as shown in (D) and (E).
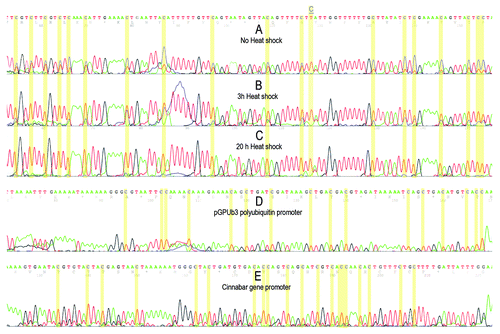
Bisulfite sequencing requires a very good control for bisulfite conversion efficiency. For example, exogenous DNA could be used as a control (e.g., by adding plasmid DNA to the genomic DNA of T. castaneum) or, as in this study, an endogenous control could be also employed. The two promoter regions from cinnabar and polyubiquitin genes of T. castaneum were chosen as controls. In addition, the same DNA used for TCAST bisulfite sequencing analysis was used to sequence the endogenous controls. The results from controls indicate a 100% of conversion after bisulfite treatment (). All experiments were repeated three times, always starting from a new preparation of genomic DNA and new oxidation by bisulfite.
The amplicon of bisulfite treated DNA from adults was also cloned and 10 clones were subsequently sequenced (). The sequences revealed a mixture of methylated and unmethylated cytosines at all positions among clones. Methylation profiles generally correspond to the profiles obtained by direct sequencing. At a single CpT position, where no methylation was detected by direct genomic sequencing, the sequencing of clones confirmed very low frequency of methylated cytosines.
Figure 6. A part (108 bp) of the most common sequence (MCS) of TCAST satellite DNA is aligned with the sequences of clones obtained after amplification of bisulfite treated DNA from adults. Each cytosine is indicated in blue and the corresponding thymines occurring when cytosines are not methylated are indicated in red. The single position where no methylation was detected by genomic bisulfite sequencing is indicated by blue column. (-) lines represent identical bases as those present in MSC of TCAST satellite DNA.

Discussion
This work describes, for the first time, cytosine methylation in the beetle Tribolium castaneum. Strong cytosine methylation was shown to be present in embryos of T. castaneum relative to larvae, pupae and adults, independently of sex. Remarkable removal of cytosine methylation during metamorphosis from embryo to larva stage was observed; the hypomethylation status continues to be present also in pupae and adults. Demethylation seems to proceed actively through 5-hydroxymethylcytosine, most probably by the action of the TET enzyme. Genome-wide epigenetic reprogramming through active and/or passive DNA demethylation in zygotes is essential for normal development in animals, but the evolutionary significance of global DNA demethylation observed in early developing embryos of most mammals remains far from clear. DNA methylation during insect embryology was till now only investigated in D. melanogaster, where DNA methylation was detected in early embryos. Most of the 5-methylcyosine is found outside of CpG dinucleotides, predominantly within CpA and CpT.Citation4 DNA methylation in D. melanogaster is catalyzed by the DNMT2 family of DNA methyltransferases and overexpression of this enzyme causes increased methylation at CpA and CpT.Citation28 Methylation at CpT and CpA is also characteristic of the stick insect Medauroidea extradentata,Citation15 while methylation of cytosines at CAA sites and CHG sites (H = A, T or C) is found in plants.Citation19,Citation29 The results presented in this study revealed that the dynamics of methylation and demethylation of the genomic DNA of T. castaneum is clearly not random. Although the level of total genomic cytosine methylation was strongly decreased from embryos to adults, the level of methylation of TCAST satellite DNA remained almost constant throughout development. Methylation within satellite DNA is not only restricted to CpG but is also found at CpA, CpT and CpC sites.
Although B. mori, D. melanogaster and T. castaneum lack DNMT3, which has the capacity to methylate DNA de novo, all three species exhibit methylation of DNA at least during specific developmental stages. This indicates that DNMT3 is not indispensable for de novo methylation and that DNMT1 and/or DNMT2 probably can provide de novo activity. What is the function of cytosine methylation in T. castaneum? The evidence of global cytosine methylation in embryos and of cytosine demethylation occurring during metamorphosis of T. castaneum indicates a possible function of this epigenetic mark in developmental regulation of insects. The constant presence of cytosine methylation and its clustering within heterochromatic major satellite DNA TCAST in all developmental stages indicates also the importance of this epigenetic modification in heterochromatin maintenance.
Heterochromatin plays an essential role in the transcriptional repression of repetitive DNA as well as in proper chromosome segregation, while DNA methylation and histone H3 lysine 9 (H3K9) methylation play a critical role in the establishment and maintenance of a repressed heterochromatin state.Citation30 Heterochromatin structure and the level of expression of heterochromatic DNA seem to be highly sensitive to environmental conditions, particularly to heat or cold stress. Heterochromatin disruption induced by heat stress during early embryogenesis in Drosophila is an epigenetic event that is transmitted to the next generation.Citation31 In plants, insects and human cells subjected to heat stress, heterochromatin-associated silencing is released and transcription of satellite DNA is significantly upregulated.Citation25,Citation32-Citation35 Activation of transcription of repetitive elements in Arabidopsis heterochromatin seems to occur due to decondensation and loss of nucleosomes,Citation34 but can be also accompanied by reduction of DNA methylation, as shown in maize.Citation36 A defect in heterochromatin DNA methylation was implicated in satellite DNA overexpression in some cancer cell lines treated with a demethylating agent (for a review see reference Citation37) or in DNMT3B-deficient human cells (for a review see reference Citation38). Heat-shock treatment of human cell lines is also associated with pericentromeric satellite DNA hypomethylation, which does not occur during treatment,Citation39,Citation40 but later in the recovery period.Citation40 However, heat shock induced hypomethylation in human cells does not regulate satellite DNA expression.Citation40 In contrast, demethylation of satellite DNA in T. castaneum occurs during heat shock treatment and depends on its lengths, as shown in this study, while activation of transcription of TCAST satellite DNA occurs during the recovery period, shortly after heat shock.Citation25 Therefore, heat stress -induced demethylation of TCAST satellite DNA seems to precede the transcriptional activation and might facilitate initiation of satellite DNA expression in T. castaneum.
In ectotherm organisms, whose body temperatures conform to ambient temperature, temperature is one of the principal environmental variables that drive adaptive evolution. In T. castaneum, upregulation of pericentromeric TCAST satellite DNA expression after heat shock is accompanied by increase in repressive epigenetic modifications of histones at TCAST regions,Citation25 and, as shown in this study, by demethylation of TCAST. Heat shock induced remodeling of heterochromatin could influence expression of genes located within heterochromatin or in its vicinity. In addition, it could result in the activation and spreading of repetitive elements within euchromatin and their insertion near genes.Citation41 Certain insertions of repetitive elements could alter gene expression and enhance adaptation under stress conditions.Citation42,Citation43 Preservation of satellite DNA methylation in all developmental stages of T. castaneum indicates the importance of this epigenetic modification for the maintenance of low transcription levels and a silent heterochromatin state. On the other hand, DNA methylation within T. castaneum constitutive heterochromatin responds to external stimuli such as heat stress. The level of demethylation depends on the duration of heat shock treatment, resulting in complete demethylation after prolonged treatment. This indicates the existence of an active DNA demethylation mechanism in T. castaneum, based probably on oxidation of methylcytosine and its further processing by specific TET enzymes. Although the biological significance of satellite DNA methylation and its link to heterochromatin remodeling is not clear, presence of an active DNA methylation/demethylation cycle in T. castaneum suggests involvement of DNA methylation in the epigenetic mechanism responsible for environmental adaptation. Further studies are necessary to elucidate the exact biological role of DNA methylation in this process.
Materials and Methods
Insect specimens
GA-2 strain of T. castaneum used for this research is obtained from Dr Dick Beeman (Manhattan, KS, USA). The same strain was used in the genome sequencing project and derives from North American wild-type strain collected in Georgia in 1982. A laboratory stock was established and maintained at a population size of > 200 individuals on standard medium (20:1, flour: brewer's yeast, by weight) in a dark incubator at 25°C and approximately 70% relative humidity.
Isolation of DNA, restriction enzymes and Southern blot hybridization
DNA was extracted from approximately 50 mg of insects in different developmental stages: embryos, larvae, pupae and adults, following the instruction of DNeasy Blood and Tissue Kit (Qiagen) for high molecular weight DNA purification. The digestion was made according to the manufacturer’s instructions, and digests were separated on 0.8% agarose gels. The methylation sensitive and non-sensitive, as well as, the methylation specific enzymes, are indicated in . The digestion was independently repeated three times on two different isolations from each developmental phases to verify the results. As a positive control of complete digestion for Hin6I, plasmid pUC18R was used as well as plasmid pHspAI2/GsaI as a control for GlaI digestion. Southern blot analyses using the biotinylated probe of the TCAST satellite DNA were performed under high stringency conditions as described previously.Citation21
Table 1. Restriction enzymes
Bisulfite conversion
Bisulfite conversion of genomic DNA (50 ng) was conducted with the MethylEasy™ DNA Bisulphite Modification Kit (Human Genetic Signatures) according to the manufacturer’s protocol. For bisulfite sequencing analysis after heat shock, adult insects were previously exposed to 40°C for 3 h and 20 h, respectively, and DNA was isolated immediately after heat shock treatment. After bisulfite treatment, DNA was immediately used for sequencing.
Bisulfite sequencing of heterochromatic TCAST satellite DNA
The methylation status of TCAST cytosines was determined by bisulfite sequencing of PCR products using methyl specific primers designed by MethPrimer software. Primers’ sequences are: forward 5′‐ATTTAAAATATATTTAATTTATTGGGTTTA‐3′ and reverse 5′‐AACTATTTTATAAAAAATTTTATCAACTTT‐3′, with expected fragment product of 265 bp. The PCR reactions were performed in a total volume of 30 μl composed of 2 × GoTaq® Green Master Mix (Promega), 2 mM mix of forward and reverse primers and 1 μl of bisulfite modified DNA. PCR conditions were as follows: 94°C for 1 min; then 10 cycles of 94°C for 30 sec, 55°C for 30 sec (-0.5°C), 70°C for 30 sec; 15 cycles of 94°C for 30 sec, 50°C for 30 sec, 70°C for 30 sec followed by a final 15 min extension at 70°C. Heat and not-heat shocked genomic DNA from GA2 population of T. castaneum was converted by bisulfite treatment: all not-methylated cytosines converted into uracils and after PCR amplification into thymine. Subsequently, PCR products purified from the gel were tested by direct sequencing to yield the overall sequence of the repeats in the genome. Each amplification product originates from a large numbers of repeats and, as expected, electropherograms show the presence of cytosine and/or thymine in the cytosine positions of the sequence.
Molecular cloning
The same PCR product used for direct bisulfite sequencing was ligated into the plasmid pGem-Teasy vector (Promega). After transformation in E. coli strain DH5a, the white colonies are directly amplified by PCR using primers T7 (TAATACGACTCACTATAGGG) and Sp6 (ATTTAGGTGACACTATAGAA). PCR reactions of 20 μl were performed using the 2x GoTaq® Green Master Mix (Promega) with the following PCR conditions: 2 min. at 95°C; 30 cycles at 94°C for 30 sec, 50°C for 30 sec and 70°C for 1 min.; 3 min. at 72°C. PCR products were analyzed by electrophoresis in a 1.3% agarose gel stained with ethidium bromide, purified using the QIAquick Gel Extraction Kit (Qiagen) and sequenced using T7 primer.
Immuno-dot blot
DNA from each T. castaneum sample as well as from standard methylated and unmethylated DNA is denatured in a 0.4 M NaOH, 1M NaCl solution and equal volumes (20 µl) of four serial dilutions (100–50–25–12 ng DNA) were spotted on two positively charged nylon membrane (Amersham Hybond-XL) with the Dot Blot 96 System (Biometra). After spotting membranes were baked for 2 h on 80°C and blocked with 1% BSA, PBT (1xPBS + 0.1% Tween-20) overnight at 4°C. After blocking, membranes were incubated with 5-mC mouse monoclonal antibody (Diagenode) following the manufacturer's protocol. For secondary antibody Biotin-Rat Anti-Mouse IgG from Invitrogen was used at a 1:10,000 dilution. The signals were detected with streptavidin-AP and BCIP/NBT alkaline phosphatase substrate (SIGMA FAST). 5-mC DNA and unmethylated DNA from the 5-mC and cytosine DNA standard pack (Diagenode) were used as positive and negative control, respectively.
Acknowledgments
This work was supported by EU FP6 Marie Curie Transfer of Knowledge Grant MTKD-CT-2006–042248, grant 00982604 from the Croatian Ministry of Science and COST Action TD0905 “Epigenetics: Bench to Bedside.” Isidoro Feliciello was a visiting scientist at Ruđer Bošković Institute.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Bird A. DNA methylation patterns and epigenetic memory. Genes Dev 2002; 16:6 - 21; http://dx.doi.org/10.1101/gad.947102; PMID: 11782440
- Feil R, Khosla S. Genomic imprinting in mammals: an interplay between chromatin and DNA methylation?. Trends Genet 1999; 15:431 - 5; http://dx.doi.org/10.1016/S0168-9525(99)01822-3; PMID: 10529801
- Suzuki MM, Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nat Rev Genet 2008; 9:465 - 76; http://dx.doi.org/10.1038/nrg2341; PMID: 18463664
- Field LM, Lyko F, Mandrioli M, Prantera G. DNA methylation in insects. Insect Mol Biol 2004; 13:109 - 15; http://dx.doi.org/10.1111/j.0962-1075.2004.00470.x; PMID: 15056357
- Xiang H, Zhu J, Chen Q, Dai F, Li X, Li M, et al. Single base-resolution methylome of the silkworm reveals a sparse epigenomic map. Nat Biotechnol 2010; 28:516 - 20; http://dx.doi.org/10.1038/nbt.1626; PMID: 20436463
- Lyko F, Foret S, Kucharski R, Wolf S, Falckenhayn C, Maleszka R. The honey bee epigenomes: differential methylation of brain DNA in queens and workers. PLoS Biol 2010; 8:e1000506; http://dx.doi.org/10.1371/journal.pbio.1000506; PMID: 21072239
- Kucharski R, Maleszka J, Foret S, Maleszka R. Nutritional control of reproductive status in honeybees via DNA methylation. Science 2008; 319:1827 - 30; http://dx.doi.org/10.1126/science.1153069; PMID: 18339900
- Wang Y, Jorda M, Jones PL, Maleszka R, Ling X, Robertson HM, et al. Functional CpG methylation system in a social insect. Science 2006; 314:645 - 7; http://dx.doi.org/10.1126/science.1135213; PMID: 17068262
- Walsh TK, Brisson JA, Robertson HM, Gordon K, Jaubert-Possamai S, Tagu D, et al. A functional DNA methylation system in the pea aphid, Acyrthosiphon pisum. Insect Mol Biol 2010; 19:Suppl 2 215 - 28; http://dx.doi.org/10.1111/j.1365-2583.2009.00974.x; PMID: 20482652
- Zemach A, McDaniel IE, Silva P, Zilberman D. Genome-wide evolutionary analysis of eukaryotic DNA methylation. Science 2010; 328:916 - 9; http://dx.doi.org/10.1126/science.1186366; PMID: 20395474
- Glastad KM, Hunt BG, Yi SV, Goodisman MAD. DNA methylation in insects: on the brink of the epigenomic era. Insect Mol Biol 2011; 20:553 - 65; http://dx.doi.org/10.1111/j.1365-2583.2011.01092.x; PMID: 21699596
- Richards S, Gibbs RA, Weinstock GM, Brown SJ, Denell R, Beeman RW, et al, Tribolium Genome Sequencing Consortium. The genome of the model beetle and pest Tribolium castaneum. Nature 2008; 452:949 - 55; http://dx.doi.org/10.1038/nature06784; PMID: 18362917
- Goll MG, Bestor TH. Eukaryotic cytosine methyltransferases. Annu Rev Biochem 2005; 74:481 - 514; http://dx.doi.org/10.1146/annurev.biochem.74.010904.153721; PMID: 15952895
- Feng S, Jacobsen SE, Reik W. Epigenetic reprogramming in plant and animal development. Science 2010; 330:622 - 7; http://dx.doi.org/10.1126/science.1190614; PMID: 21030646
- Krauss V, Eisenhardt C, Unger T. The genome of the stick insect Medauroidea extradentata is strongly methylated within genes and repetitive DNA. PLoS One 2009; 4:e7223; http://dx.doi.org/10.1371/journal.pone.0007223; PMID: 19787064
- Mandrioli M. Cytogenetic characterization of telomeres in the holocentric chromosomes of the lepidopteran Mamestra brassicae.. Chromosome Res 2002; 10:279 - 86; http://dx.doi.org/10.1023/A:1016515607278; PMID: 12199141
- Mandrioli M, Volpi N. The genome of the lepidopteran Mamestra brassicae has a vertebrate-like content of methyl-cytosine. Genetica 2003; 119:187 - 91; http://dx.doi.org/10.1023/A:1026016021415; PMID: 14620958
- Mandrioli M, Azzoni P, Lombardo G, Manicardi GC. Composition and epigenetic markers of heterochromatin in the aphid Aphis nerii (Hemiptera: Aphididae). Cytogenet Genome Res 2011; 133:67 - 77; http://dx.doi.org/10.1159/000323510; PMID: 21273762
- Lyko F, Maleszka R. Insects as innovative models for functional studies of DNA methylation. Trends Genet 2011; 27:127 - 31; http://dx.doi.org/10.1016/j.tig.2011.01.003; PMID: 21288591
- Kangaspeska S, Stride B, Métivier R, Polycarpou-Schwarz M, Ibberson D, Carmouche RP, et al. Transient cyclical methylation of promoter DNA. Nature 2008; 452:112 - 5; http://dx.doi.org/10.1038/nature06640; PMID: 18322535
- Métivier R, Gallais R, Tiffoche C, Le Péron C, Jurkowska RZ, Carmouche RP, et al. Cyclical DNA methylation of a transcriptionally active promoter. Nature 2008; 452:45 - 50; http://dx.doi.org/10.1038/nature06544; PMID: 18322525
- Feliciello I, Chinali G, Ugarković Đ. Structure and population dynamics of the major satellite DNA in the red flour beetle Tribolium castaneum. Genetica 2011; 139:999 - 1008; http://dx.doi.org/10.1007/s10709-011-9601-1; PMID: 21837441
- Ito S, D’Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature 2010; 466:1129 - 33; http://dx.doi.org/10.1038/nature09303; PMID: 20639862
- Ugarković Đ, Podnar M, Plohl M. Satellite DNA of the red flour beetle Tribolium castaneum--comparative study of satellites from the genus Tribolium.. Mol Biol Evol 1996; 13:1059 - 66; http://dx.doi.org/10.1093/oxfordjournals.molbev.a025668; PMID: 8865660
- Pezer Ž, Ugarković Đ. Satellite DNA-associated siRNAs as mediators of heat shock response in insects. RNA Biol 2012; 9:587 - 95; http://dx.doi.org/10.4161/rna.20019; PMID: 22647527
- Feliciello I, Picariello O, Chinali G. The first characterisation of the overall variability of repetitive units in a species reveals unexpected features of satellite DNA. Gene 2005; 349:153 - 64; http://dx.doi.org/10.1016/j.gene.2004.12.001; PMID: 15777738
- Feliciello I, Picariello O, Chinali G. Intra-specific variability and unusual organization of the repetitive units in a satellite DNA from Rana dalmatina: molecular evidence of a new mechanism of DNA repair acting on satellite DNA. Gene 2006; 383:81 - 92; http://dx.doi.org/10.1016/j.gene.2006.07.016; PMID: 16956734
- Kunert N, Marhold J, Stanke J, Stach D, Lyko FA. A Dnmt2-like protein mediates DNA methylation in Drosophila. Development 2003; 130:5083 - 90; http://dx.doi.org/10.1242/dev.00716; PMID: 12944428
- Zakrzewski F, Weisshaar B, Fuchs J, Bannack E, Minoche AE, Dohm JC, et al. Epigenetic profiling of heterochromatic satellite DNA. Chromosoma 2011; 120:409 - 22; http://dx.doi.org/10.1007/s00412-011-0325-x; PMID: 21594600
- Lippman Z, Martienssen R. The role of RNA interference in heterochromatic silencing. Nature 2004; 431:364 - 70; http://dx.doi.org/10.1038/nature02875; PMID: 15372044
- Seong KH, Li D, Shimizu H, Nakamura R, Ishii S. Inheritance of stress-induced, ATF-2-dependent epigenetic change. Cell 2011; 145:1049 - 61; http://dx.doi.org/10.1016/j.cell.2011.05.029; PMID: 21703449
- Jolly C, Metz A, Govin J, Vigneron M, Turner BM, Khochbin S, et al. Stress-induced transcription of satellite III repeats. J Cell Biol 2004; 164:25 - 33; http://dx.doi.org/10.1083/jcb.200306104; PMID: 14699086
- Valgardsdottir R, Chiodi I, Giordano M, Cobianchi F, Riva S, Biamonti G. Structural and functional characterization of noncoding repetitive RNAs transcribed in stressed human cells. Mol Biol Cell 2005; 16:2597 - 604; http://dx.doi.org/10.1091/mbc.E04-12-1078; PMID: 15788562
- Pecinka A, Dinh HQ, Baubec T, Rosa M, Lettner N, Mittelsten Scheid O. Epigenetic regulation of repetitive elements is attenuated by prolonged heat stress in Arabidopsis. Plant Cell 2010; 22:3118 - 29; http://dx.doi.org/10.1105/tpc.110.078493; PMID: 20876829
- Pezer Ž, Brajković J, Feliciello I, Ugarković Đ. Transcription of Satellite DNAs in Insects. Prog Mol Subcell Biol 2011; 51:161 - 78; http://dx.doi.org/10.1007/978-3-642-16502-3_8; PMID: 21287138
- Hu Y, Zhang L, He S, Huang M, Tan J, Zhao L, et al. Cold stress selectively unsilences tandem repeats in heterochromatin associated with accumulation of H3K9ac. Plant Cell Environ 2012; 35:2130 - 42; http://dx.doi.org/10.1111/j.1365-3040.2012.02541.x; PMID: 22616903
- Eymery A, Callanan M, Vourc’h C. The secret message of heterochromatin: new insights into the mechanisms and function of centromeric and pericentric repeat sequence transcription. Int J Dev Biol 2009; 53:259 - 68; http://dx.doi.org/10.1387/ijdb.082673ae; PMID: 19412885
- Hall LE, Mitchell SE, O’Neill RJ. Pericentric and centromeric transcription: a perfect balance required. Chromosome Res 2012; 20:535 - 46; http://dx.doi.org/10.1007/s10577-012-9297-9; PMID: 22760449
- Eymery A, Horard B, El Atifi-Borel M, Fourel G, Berger F, Vitte AL, et al. A transcriptomic analysis of human centromeric and pericentric sequences in normal and tumor cells. Nucleic Acids Res 2009; 37:6340 - 54; http://dx.doi.org/10.1093/nar/gkp639; PMID: 19720732
- Tilman G, Arnoult N, Lenglez S, Van Beneden A, Loriot A, De Smet C, et al. Cancer-linked satellite 2 DNA hypomethylation does not regulate Sat2 non-coding RNA expression and is initiated by heat shock pathway activation. Epigenetics 2012; 7:903 - 13; http://dx.doi.org/10.4161/epi.21107; PMID: 22722874
- Brajković J, Feliciello I, Bruvo-Mađarić B, Ugarković Đ. Satellite DNA-like elements associated with genes within euchromatin of the beetle Tribolium castaneum. G3: Genes, Genomes. Genetics 2012; 2:931 - 41
- Kalendar R, Tanskanen J, Immonen S, Nevo E, Schulman AH. Genome evolution of wild barley (Hordeum spontaneum) by BARE-1 retrotransposon dynamics in response to sharp microclimatic divergence. Proc Natl Acad Sci U S A 2000; 97:6603 - 7; http://dx.doi.org/10.1073/pnas.110587497; PMID: 10823912
- Pezer Ž, Brajković J, Feliciello I, Ugarković D. Satellite DNA-mediated effects on genome regulation. Genome Dyn 2012; 7:153 - 69; http://dx.doi.org/10.1159/000337116; PMID: 22759818