Abstract
Overexpression of EZH2 and other PRC2 subunits, such as SUZ12, is associated with tumor progression and poor prognosis in several human malignancies. Nevertheless, the underlying mechanisms driving aberrant EZH2 expression are poorly understood. This review provides molecular insights into the essential role of EZH2 in breast and prostate tumorigenesis. We addressed the current understanding on the oncogenic role of EZH2, with an emphasis on: (1) the less known PRC2-independent role of EZH2 in gene activation, in addition to its canonical role in transcriptional silencing as a histone methyltransferase catalyzing the trimethylation of histone H3 at lysine 27; (2) causes and consequences of its deregulation in tumor cells and; (3) collaboration of EZH2 with other epigenetic and hormone receptor-mediated oncogenic signaling pathways. We also summarize how EZH2 has emerged as a promising therapeutic target in hormone-refractory cancers and the prospects for integrating EZH2 blockade with available pharmacological inhibitors.
Introduction
Moving ahead from the conventional notion of “Oncogenesis” as a complex series of events driven by the accumulation of genetic mutations,Citation1 technological advances in recent years have highlighted that the collusion between genetic and epigenetic regulatory mechanisms is prevalent and its disruption contributes significantly to the initiation and progression of cancer.Citation2 This crosstalk is being established by genetic mutations in several epigenetic modifiers that are involved in DNA and histone modifications, leading to abnormal epigenetic alterations that cause aberrant gene expression and genomic instability, ultimately resulting in cancer.Citation3,Citation4
In addition to epigenetic aberration, such as promoter DNA hypermethylation, two discrete histone modifications, i.e., histone H3 lysine 27 trimethylation (H3K27me3) and H3K9 dimethylation (H3K9me2), catalyzed by histone lysine methyltransferases, significantly contribute to epigenetic silencing mechanism in mammals.Citation5-Citation9 H3K27me3-mediated gene silencing involves an evolutionarily conserved family of chromatin modulators called Polycomb group (PcG) proteins.Citation10 PcG proteins are best known for their role in maintaining the silenced state of several genes during development and establishing embryonic stem cell (ESC) identity and pluripotency.Citation11 Enhancer of zeste homolog 2 (EZH2), the catalytic subunit of the Polycomb repressive complex 2 (PRC2), trimethylates H3K27, which may serve as a docking site for another Polycomb repressor complex, PRC1, as well as for DNA methyltransferases (DNMTs).Citation10,Citation12 Besides promoter DNA methylation, EZH2-mediated H3K27me3 is a potential independent mechanism of epigenetic silencing of tumor suppressor genes in cancer.Citation13,Citation14
Recent findings implicate that EZH2 is overexpressed in a wide range of cancer types, especially breast and prostate cancers.Citation15,Citation16 Breast and prostate cancer are the two most well characterized endocrine-related cancers that share defining similarities in the key hormone signaling pathways controlling tumor growth and differentiation.Citation17 In this review, we comprehensively discuss the molecular mechanism of action of EZH2 and how its deregulation leads to tumorigenesis in these endocrine-related cancers. Emerging literature suggests that EZH2 functions as a multifaceted molecule and switches to the role of a transcriptional activator inducing a subset of genes that promote oncogenesis in certain cancer types, such as castration-resistant prostate cancer (CRPC) and breast cancer.Citation18-Citation21 Recent literature highlighting the relationship between EZH2-mediated gene silencing and other epigenetic silencing mechanisms, i.e., DNA methylation and histone modifications, are discussed in detail. We also highlight some important studies which link EZH2 with estrogen- and androgen-receptor-mediated signaling in breast and prostate tumorigenesis, respectively. We contend that EZH2 may emerge as a potential therapeutic target in hormone-refractory breast and prostate cancers and in the development of novel treatment strategies that target both genetic and epigenetic mechanisms of tumorigenesis. This may help combat the issue of hormone resistance in current treatment regimens.
Polycomb Group (PcG) Proteins: Composition, Recruitment and Action
Polycomb group proteins (PcG) are a phylogenetically conserved group of multimeric protein complexes, initially characterized in Drosophila for their crucial role as negative regulators of homeotic genes required for segmentation.Citation22 In mammals, PcG proteins function in two distinct protein complexes: Polycomb repressive complexes PRC1 and PRC2. Posttranslational modification of histones has emerged as the key molecular mechanism of action of PcG proteins, which regulate chromatin structure and mediate epigenetic silencing. PRC1 composition is variable and its core components in mammals include Bmi1 (also known as Pcgf4), Ring1b, Ring1a, CBX, PH1, PH2, NSPC1 (Pcgf1) and MEL18 (Pcgf2) proteins.Citation23,Citation24 Ring 1a and Ring 1b ubiquitin ligases catalyze the monoubiquitination of histone 2A at lysine 119 (H2AK119ub), which causes PRC1 dependent gene silencing.Citation25 ncRNA and H3K27me3 facilitate PRC1 recruitment to target genes.Citation26 It blocks the ATP-dependent remodeling activity of Swi/Snf in vitro and maintain the repressed state of genes.Citation27
The mammalian PRC2 core complex consists of four components: EZH1/2, SUZ12, EED and RbAp46/48 (also called RBBP7/4) that catalyzes the di/trimethylation of H3K27.Citation10,Citation28 Recent reports suggest that there are additional PRC2 components, AEBP2, PCLs and JARID2, which function as accessory units regulating the function and enzymatic activity of PRC2 holoenzyme.Citation29-Citation31 Despite the dynamic composition of PRC2, EZH1/2 remains its integral component for catalyzing H3K27 di/trimethylation.Citation32 EZH1 and EZH2 are two paralogs and PRC2-EZH1/-EZH2 complexes function differently to maintain the repressed state of chromatin.Citation28 EZH1 expression has been reported in dividing and differentiated cells whereas EZH2 is found in proliferating tissues.Citation28,Citation33 Since both are part of PRC2 complexes, they regulate an overlapping array of target genes. Intriguingly, PRC2-EZH1 has low histone methyltransferase (HMTase) compared with EZH2-containing PRC2 complex, which suggests that the H3K27me3 repressive mark is established by PRC2-EZH2.Citation28 HMTase activity of PRC2-EZH1 may function in restoring the repressive methylation pattern (H3K27me2/me3) of histones that could have lost their methylation mark after demethylase activity or histone exchange.
Human EZH2 gene maps to the long arm of chromosome 7 at position 7q35 and encodes a 746 amino acid protein that belongs to the histone-lysine methyltransferase family.Citation34 EZH2 protein contains several functional domains that are involved in mediating its interaction with other PRC2 and regulatory proteins, besides its histone methyltransferase activity. The major domains include CXC (cysteine-rich domain), SET (Su(var)3–9, enhancer of zeste, trithorax domain), ncRBD (non-coding RNA-binding domain and a DNA binding domain.Citation35 Although mutations in other PRC2 members have not been reported so far, recent studies in lymphoma and myeloid neoplasms have identified EZH2 mutations that may either cause gain-of-function or complete loss of histone methyltransferase activity.Citation36 A heterozygous missense somatic mutation at tyrosine 641 in the SET domain of EZH2 results in enhanced H3K27me3 levels in follicular lymphomas and diffused large B-cell lymphomas.Citation37 Interestingly, in myeloid neoplasms, inactivating EZH2 mutations that are distributed throughout the gene, comprising missense, nonsense and premature stop codons, have been described and lead to loss of histone methyltransferase activity.Citation38,Citation39
EZH2 protein is subjected to various posttranslational modifications, such as phosphorylation by Akt1 at serine 21, which reduces H3K27 trimethylation activity and phosphorylation at threonine 345 by CDK1 and CDK2, which is required for maintenance of H3K27me3 repressive marks at target gene promoters.Citation40,Citation41 Furthermore, a recent study demonstrates that EZH2 and SUZ12 are potential targets for sumoylation in both in vivo and in vitro conditions.Citation42
In Drosophila, PcG proteins are recruited to polycomb response elements (PREs) containing target genes with the help of several other DNA binding proteins such as GAF, Pipsqueak, Zeste or PHO. In mammals, PRC2 is targeted to CpG islands but, surprisingly, no consensus sequence elements for PcG recruitment have been identified to date in this species.Citation35 Recent reports suggest that YY1, the mammalian ortholog of the Drosophila PRE DNA-binding protein PHO and RYBP, which interacts with both PcG proteins, is involved in the PRC2 recruitment process.Citation43-Citation45 Long non-coding RNAs (ncRNA) have also emerged as potential elements involved in PRC2 recruitment. HOTAIR ncRNA promote PRC2 recruitment in trans and is associated with the transcriptional repression of the HOXD locus.Citation46,Citation47 Similarly, XIST and KCNQ1OT1 ncRNAs have also been implicated in PRC2 gene targeting.Citation48-Citation51
There are two possible molecular mechanisms for EZH2 action based on its role as a transcriptional activator or repressor. As an integral component of PRC2, the canonical role of EZH2 is that of a histone methyltransferase (). The SET domain of EZH2 catalyzes methylation of lysine in succession so that each methylation event serves as a substrate for the next (H3K27me2 is monomethylated to form H3K27me3); each methylation mark represents functionally distinct chromatin state. H3K27me3 has been implicated in the recruitment of PRC1 complex, suggesting that both PcG protein complexes function in gene silencing. However, there are PRC2 target genes that lack H2AK119ub and genes targeted by PRC1 in the absence of PRC2, highlighting the discrepancy in the exact functional relationship between the two protein complexes.Citation43,Citation52,Citation53 Trimethylation of H3 may pose steric hindrance for RNAP II and other proteins binding to target gene promoters and repress transcription.
Figure 1. Molecular mechanism of action of EZH2. (A) Polycomb dependent mechanism—Role in transcriptional repression. EZH2 functions as a part of mammalian PRC2 core complex consisting of EZH1/2, SUZ12, EED and RbAp46/48 (also called RBBP7/4. When recruited to the target gene promoter it catalyses the di/tri- methylation of Histone 3 at lysine 27 (H3K27me3), resulting in chromatin compaction and inaccessibility of promoter region to RNA Pol II and other proteins of the transcription machinery, which ultimately repress transcription. H3K27me3 mark also serves as a docking site for binding of PRC1 complex containing Bmi1, Ring1a, Ring1b, HPH1, HPH2, NSPC1, MEL18 and CBX proteins (-2, 4, 6, 7, 8). PRC1 catalysed mono-ubiquitination of Histone 2A at lysine 119 further contributes to target gene silencing. (B) Polycomb independent mechanism—Role in transcriptional activation. (1) In prostate cancer cells, Akt-1 mediated phosphorylation of EZH2 at Serine-21 decreases the H3K27me3 activity of EZH2. But phosphorylated EZH2 can function independent of other PRC2 proteins and may methylate androgen receptor at lysine 630 and 632, which can enhance its transcriptional activity. This is a potential mechanism for EZH2 mediated transcriptional activation via methylation of androgen receptor or other androgen receptor associated proteins (X or Y). (2) When over-expressed in ER-positive, luminal like MCF-7 breast cancer cells, EZH2 functions as a transcriptional activator by acting as a bridge to physically link ERα and Wnt signaling components β-catenin and TCF, on the Cyclin B1 and c-Myc promoters. EZH2 also associates with Mediator complex through its domain II independent of the SET domain involved in HMTase activity and enhance transcription by its interaction with RNA polymerase II. In ER-negative, basal like MDA-MB-231 cells, EZH2 forms a ternary complex with NF-kB components RelA and RelB and activates transcription of NF- kB target genes such as TNF, IL6.
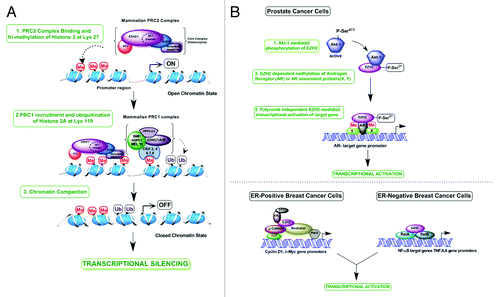
The second mechanism of EZH2 action highlights its less known role as a transcriptional inducer. Xu et al.Citation18 reported that in CRPC, EZH2 functions independently of other PRC2 members and Akt1-mediated phosphorylation of EZH2 at serine 21 allows it to interact with AR at many solo genes (). They also demonstrated that depletion of EZH2 does not alter AR levels but induces a reduction in AR-associated lysine methylation (lysines 630 and 632). AR methylation catalyzed by Set9 enhances its transcriptional activity.Citation54,Citation55 Therefore, methylation of AR or AR-associated proteins is a potential mechanism for EZH2-driven transcriptional activation.
In breast cancer cells, EZH2 has been reported to induce gene transcription in two different ways depending on the ER status (). In luminal-like MCF-7 breast cancer cells, EZH2 acts as a bridge to physically link ERα and Wnt signaling components β-catenin and TCF, on the cyclin B1 and c-Myc promoters.Citation21 EZH2 also interacts with Mediator complex through its domain II independent of the SET domain involved in HMTase activity and induces transcription by its interaction with RNAP II. In ER-negative, basal like MDA-MB-231 cells, EZH2 forms a ternary complex with NF-κB components RelA and RelB and activates transcription of NF-κB target genes such as TNF and IL6.Citation20 In both types of cancer, although EZH2 functions independently of PRC2 as a transcriptional activator, the mechanisms are different.
H3K27me3 and DNA methylation
Epigenetic modulations such as aberrant promoter DNA hypermethylation and histone modifications, which govern chromatin remodeling, have emerged as important players in tumor onset and progression. Interesting experimental studies addresses the functional relationship between PcG proteins and DNA methylation that leads to epigenetic inactivation of tumor suppressor genes in various cancer types. Previous studies show that the crosstalk between DNA methylation and histone modifications is mediated by a group of proteins possessing methyl DNA binding characteristics such as methyl CpG binding protein 2 (MeCP2), methyl CpG binding domain protein 1 (MBD1) and Kaiso/ ZBTB 33 (Zinc finger and BTB domain containing protein 33).Citation56,Citation57 These proteins bind to methylated CpG islands on target gene promoters and recruit histone deacetylase (HDAC) and histone methyltransferase-containing protein complexes, adding further to target gene silencing. But the evidences linking DNA methylation and PRC-mediated silencing are partial and sometimes conflicting. Recent technological developments have led to multiple mechanistic hypotheses on the underlying process.
Early studies in Drosophila and C. elegans suggest the involvement of PRC2-mediated repressive machinery in gene silencing but no detectable DNA methylation in the chromatin.Citation58 Vire et al.Citation59 first reported the role of EZH2 and its H3K27me3 repressive mark in the recruitment of DNMTs to EZH2 target promoter in human osteosarcoma cells. They described that EZH2 is essential for the binding of DNMT, but DNMT is not required for EZH2 or H3K27me3 binding to the MYT1 gene promoter. Bisulfite genomic sequencing data of MYT1 and WNT1 promoters in EZH2 overexpressing and knockdown cells show that EZH2 is required for CpG methylation of EZH2 target promoters. McGarvey et al.Citation60 demonstrated that EZH2 might act as an epigenetic modulator when promoter is not densely methylated. They showed that EZH2 knockdown lead to increased expression of unmethylated or basally expressing genes but not hypermethylated completely silenced genes such as p16INK4a in U2OS cells. Contrary to previous reports, they found that EZH2 depletion does not reduce promoter DNA methylation of target genes and proposed a dominant role for dense CpG island DNA methylation in maintaining the heritable repressive state of target genes as compared with various other repressive chromatin marks.
To further address the issue of PcG proteins recruiting DNMTs and establish a functional link between them, Kondo et al.Citation13 analyzed DNA methylation at several H3K27me3 modified CpG islands in human prostate cancer cell line PC3 and breast cancer cell line MCF-7. Using bisulfite pyrosequencing in 2–6 adjacent CpG sites in PC3 cells, they confirmed that methylation was very low in candidate genes, ranging from 6.3% to 39.3%, with only three out of eight genes having methylation above 15%. Such low levels of DNA methylation compared with what has been generally observed, is insufficient for causing silencing of tumor suppressor genes. Global assessment of DNA methylation and H3K27me3 in PC3 and MCF-7 cells focusing on genes detectable on both ChIP-chip and DNA methylation arrays, using methylated CpG island microarrays (MCAM), revealed that H3K27-based silencing and DNA methylation are independent events. However, there was some overlap between H3K27me3 and DNA methylation, which suggest that there may be genes targeted by both silencing mechanisms, although it seems relatively rare.
Absence of DNA hypermethylation at PRC-occupied regions in embryonic carcinoma cells and lack of DNA hypomethylation after EZH2 knockdown in cancer cells hint at different gene sets being targeted for repression by these two silencing machineries. However, some gene profile studies comparing chromatin states in tumor cells vs. cancer cells revealed that H3K27me3-enriched EZH2 target genes in normal cells subsequently become abnormally hypermethylated in tumor cells during cellular transformation.Citation61-Citation63 In conclusion, the current view supported by comprehensive genome-wide and functional analysis suggests that DNA methylation and Polycomb-mediated silencing collaborate in the epigenetic silencing of certain genes depending on cellular context but are probably not as directly related as previously suggested by some reports.
H3K27me3 and other histone modifications
Histone modification is a well-conserved and important mechanism of transcriptional regulation. Emerging evidence suggests that the modification patterns of histones can be linked to their biological function and act as a “histone code,” to predict the transcriptional states of genes. Histone methylation at H3K4 (trimethylation), enriched at promoter/transcription start site regions, and H3K36 (dimethylation), most abundant in 5′ coding regions, are hallmarks of actively transcribed genes.Citation64-Citation66 Many studies comprehensively described H3K4me3 and H3K36me2 antagonizing Polycomb-mediated silencing at H3K27.Citation67,Citation68 This inhibition requires the presence of these methylation marks on the same histone tail subject to EZH2-mediated K27 methylation. Similarly, H3K27 acetylation and H3K27me3 are mutually exclusive and, hence, acetylation of H3K27 functionally antagonizes PRC2-mediated silencing in both mammalian and fly systems.Citation69-Citation71 This highlights the role of HDACs in EZH2-mediated silencing, although they are not core subunits of PRC2. HDACs could deacetylate K27 side chains to make ɛ-amino groups available for methylation by the PRC2 complex. Also, two recent studies establish that phosphorylation of serine 28, located immediately next to H3K27, by kinases MSK1 and MSK2 also disrupts the functioning of PcG silencing pathway.Citation72,Citation73
In summary, recent literature advocates the fact that PRC2-mediated gene silencing mechanisms have evolved to sense the surrounding epigenetic landscape and are subject to cues from other chromatin marks based on the transcriptional state of the chromatin.Citation74 However, further studies addressing the structural organization of PRC2 active site and its regulatory domains are needed to completely unravel its mechanism of inhibition.
EZH2 and endocrine-Related Cancers
EZH2 in breast tumorigenesis
Breast cancer is the most common malignancy and the second leading cause of cancer-related deaths, accounting for nearly one in three cancers diagnosed among women in the United States. According to recent breast cancer statistics, approximately 230,480 new cases of invasive breast cancer and 39,520 breast cancer deaths among US women were reported in 2011.Citation75 Breast cancer is a heterogeneous malignancy and can be categorized into different subtypes based on molecular profiling. Each subtype varies in their response to therapy directed against them and overall survival, highlighting the need for a more specific and personalized treatment regimen. Despite recent advances in molecular cancer research, only few biomarkers of breast cancer progression have been proven to be clinically useful, such as estrogen receptor (ER), progesterone receptor (PR), erbB2 overexpression, uPAR and Cathepsin D, among others.Citation76,Citation77 Current treatment strategies include drugs such as herceptin and tamoxifen or aromatase inhibitors for erbB2-overexpressing tumors and ER-positive tumors, respectively. Chemotherapy is the standard treatment for tumors that are poorly differentiated and lack erbB2, ER and PR. More effective therapeutic strategies and reliable prognostic markers are therefore needed to improve the clinical outcome of patients with breast cancer.Citation78
Metastasis is responsible for the majority of cancer-related deaths and the invasiveness or metastatic potential of breast cancer is inversely correlated with the degree of tumor differentiation. Highly undifferentiated tumors represented clinically by nuclear pleomorphism and the formation of glandular structures, have greater probability to develop metastasis. Several studies proposed EZH2 as a promising novel biomarker for aggressive breast cancer associated with poor prognosis.Citation15,Citation79,Citation80 EZH2 protein levels have been reported to increase steadily through successive stages of neoplastic transformation from normal epithelium to epithelial hyperplasia, ductal carcinoma in situ, invasive carcinoma and distant metastasis. In fact, EZH2 has emerged as an independent predictor of recurrence and death in patients with breast cancer.
Molecular insights into EZH2-mediated breast tumorigenesis
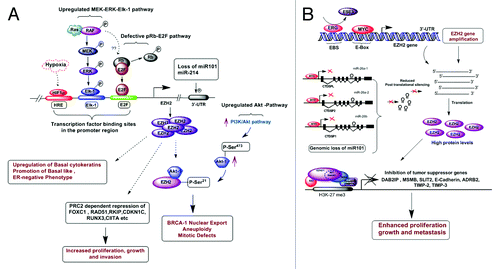
Several experimental studies have established that elevated EZH2 levels in human breast carcinomas is associated with the aggressive ER-negative basal-like phenotype characterized by lack of ER expression, nuclear polymorphism and lack of BRCA1 protein.Citation15,Citation79-Citation81 Several essential tumor suppressor genes are repressed, ultimately leading to metastasis in breast carcinomas ().Citation20,Citation82-Citation88 Deregulation of EZH2 contributes to ER-negative breast cancer progression. EZH2 knockdown in breast cancer cells caused decreased proliferation and delayed the G2/M cell-cycle transition. In vivo studies demonstrated that EZH2 downregulation significantly decreased breast xenograft growth and improved survival.Citation81 EZH2 knockdown upregulated BRCA1, a nuclear protein that plays a major role in DNA repair and estrogen receptor modulation.Citation89 The proposed mechanisms underlying EZH2 overexpression, which ultimately causes uncontrolled cell proliferation leading to tumorigenesis in breast, is discussed in the following section ().
Table 1. Some metastasis associated target genes epigenetically repressed by EZH2 in breast and prostate cancer
Figure 2. Molecular insights into EZH2 driven breast and prostate tumorigenesis. (A) In breast cancer cells EZH2 expression is regulated by several factors such as hypoxia induced HIFα, pRB-E2F and MEK-ERK-Elk1 pathways. Genomic loss of miR101 and miR214 also up-regulates EZH2 expression. Elevated EZH2 levels leads to the transcriptional repression of several tumor suppressor genes such as FOXC1, RAD51, RKIP, CDKIC, RUNX3, CIITA etc, by PRC2 mediated H3K27 trimethylation. High EZH2 protein levels are associated with increased expression of phospho-Akt1 (Ser473) and decreased nuclear localization of phospho-BRCA1 (Ser1423). EZH2 mediated nuclear shuttling of BRCA-1 protein in ER negative basal like breast cancer cells is one of its PRC2 independent functions (others described in ). Nuclear retention of BRCA-1 protein leads to aneuploidy, aberrant mitosis and genomic instability, which ultimately promotes tumorigenesis. (B) In prostate cancer cells, four molecular mechanisms are reported to be responsible for EZH2-amplification or overexpression of the EZH2 gene including deletion of its negative regulator miR-101, transcriptional regulation by MYC and ETS gene family members. MYC binds upstream of EZH2 promoter and induces EZH2 expression. Interestingly MYC represses the transcription of CTDSPL, CTDSP2 and CTDSP1 which harbor miR-26a and miR-26b. Repression of miR-26a and miR-26b contribute additionally to EZH2 overexpression as miR-26a and miR-26b would be unavailable to destabilize EZH2 mRNA by binding specifically to the EZH2 3'-UTR in RISC complex. Furthermore, ETS transcriptional network also regulates the expression of EZH2. Epithelial-specific ETS factor ESE3 represses EZH2 expression whereas ERG, binds to the promoter of EZH2 and competes with ESE3 for promoter occupancy opposing its effects. EZH2 overexpression leads to H3K27 methylation associated silencing of critical tumor suppressor genes such as DAB2IP, MSMB, SLIT, TIMP-2, and TIMP-3, which contribute to increased growth, proliferation and invasive phenotype of prostate cancer cells.
EZH2 overexpression in breast cancer: causes and consequences
pRB-E2F signaling pathway
The pRB-E2F pathway plays a key role in regulating mammalian cell cycle progression. Defects in the core members of this pathway are frequently reported in different malignancies and, hence, inactivation of pRB-E2F pathway has emerged as a mandatory event in all human cancers.Citation1 The transcription factors from the E2F family are well-studied targets of pRB and regulate genes that control entry into the S-phase and DNA replication, such as cyclinE1, cyclinA2, CDC6, DHFR and TK1.Citation90-Citation92 Bracken et al.Citation93 reported two interesting findings that present a convincing direct link between pRB-E2F pathway and PRC-mediated histone modifications. First, PRC2 components EZH2 and EED are essential for cell proliferation and regulate the expression of “positive regulators” of cell cycle such as cyclinD1 (CCND1), cyclinE1 (CCNE1), cyclinA2 (CCNA2) and cyclinB1 (CCNB1). Interestingly, abrogation of EZH2 or EED expression did not upregulate the expression of negative regulators of the cell cycle; however, the expression of several E2F-regulated genes, such as G1/S cyclin, was significantly decreased. Second, they showed that E2Fs transactivate and bind to EZH2 or EED promoters, suggesting that PRC2-mediated cell cycle control occurs downstream of the pRB-E2F pathway.
MEK-ERK1/2-Elk-1 pathway
The MEK-ERK-Elk-1 pathway is reported to be frequently upregulated in various cancer types. Fujii et al. have shown that the MEK-ERK-Elk-1 pathway is linked to EZH2 overexpression in breast cancer cells.Citation94 EZH2 promoter analysis results revealed three Elk-1 binding motifs as well as other response elements such as NF-κB, c-Myb, STAT1 and SRF (serum response factor), which cooperate with Elk-1 for transcriptional activation. It was demonstrated that MEK inhibitor treatment and Elk-1 knockdown leads to decreased phospho-Elk-1 binding to the EZH2 promoter. In triple negative/basal type breast cancer cells, the MEK/ERK pathway activated via KRAS mutation, ERBB2 amplification and EGFR amplification contributes to EZH2 overexpression.
Hypoxia-induced EZH2 overexpression
In breast tumor initiating cells (BTICs), using promoter analysis, Chang et al.Citation95 identified a consensus sequence for HIF response element (HRE) within the EZH2 promoter region. The authors reported that HIF transcription factor (HIF1α) binds to the HRE-containing EZH2 promoter region in a hypoxic microenvironment and transactivates EZH2 expression. Increased EZH2 expression was found to be inversely related to double-strand break repair protein RAD51 expression, which ultimately causes genomic instability due to impaired DNA damage repair. RAF1 gene amplification was also reported in BTICs due to EZH2-mediated downregulation of DNA repair, which further activates p-ERK-β-catenin signaling. The overall functional effect is enhanced self-renewal and expansion of BTIC population, ultimately facilitating breast cancer progression.
Akt-mediated regulation
Posttranslational modification plays a critical role in regulating EZH2 levels. Cha et al.Citation40 reported that breast cancer cells with elevated levels of phosphorylated Akt have lower levels of H3K27me3. EZH2 protein has one highly conserved, potential Akt phosphorylation site at serine 21. Akt phosphorylates EZH2 at serine 21, which inhibits K27 methylation of histone H3. This event has no effect on the composition of the PRC2 complex, but it reportedly reduces the affinity of EZH2 toward histone H3 causing decreased H3K27 trimethylation and de-repression of normally silenced EZH2 target genes.
EZH2, BRCA1 and PI3K/Akt crosstalk
BRCA1 is a tumor suppressor gene that encodes a nuclear protein involved in numerous functions, such as DNA repair, activation of cell cycle checkpoints and maintenance of chromosome stability.Citation96,Citation97 Women with an inherited mutation in one BRCA1 allele develop basal-like breast tumors when there is loss of function of the second BRCA1 allele and concomitant loss of p53.Citation98 BRCA1-deficient breast tumors show undifferentiated basal like phenotype and aggressive behavior.Citation99-Citation101 Recent reports suggest that elevated EZH2 levels in BRCA1-deficient human breast tumors and are associated with poor survival.Citation15,Citation79,Citation80 Mechanistically, upregulation of EZH2 in benign mammary epithelial cells inhibits BRCA1 phosphorylation (serine 1423) and causes nuclear export of BRCA1 protein with high levels of Cdc2–CyclinB1 complex (essential for G2/M checkpoint control), ultimately resulting in uncontrolled cell division, aberrant mitoses with extra chromosomes and genomic instability.Citation89 Conversely, both in vivo and in vitro studies confirm that EZH2 inhibition increases the protein levels of total BRCA1 and of BRCA1 phosphorylation at serine 1423 and induces BRCA1 nuclear localization, which restores defects in mitosis and ploidy level. Gonzalez et al.Citation89 have demonstrated that EZH2 overexpression in breast cancer cells causes activation of the phosphoinositide-3-kinase/Akt (PI3K/Akt) pathway, specifically through activation of the Akt isoform. In 39% of invasive breast carcinomas, high EZH2 protein levels are associated with increased expression of phospho-Akt-1 (serine 473) and decreased nuclear localization of phospho-BRCA1 (serine 1423). EZH2 requires Akt1 to regulate the intracellular localization of BRCA1. Therefore, it may be inferred that Akt1-mediated phosphorylation of EZH2 (serine 21) regulates the shuttling of BRCA1 between nucleus and cytoplasm.
EZH2 and estrogen receptor signaling
The role of estrogen signaling, either via canonical or genomic non-classical pathways, is well established in breast carcinogenesis. Emerging literature suggests that epigenetic factors play an instrumental role in regulating estrogen signaling in breast cancer.Citation102 As discussed in previous sections, several in vivo and in vitro studies have confirmed that high EZH2 levels correlate with malignant and highly aggressive breast tumors. Therefore, it is logical to investigate the connection between estrogen signaling and EZH2 activity in breast carcinoma cells. In most of its biological roles, EZH2 mainly functions as a transcriptional repressor; however, in breast cancer cells, it transactivates genes commonly targeted by estrogen and Wnt signaling pathways. Shi et al.Citation21 demonstrated that Polycomb group protein EZH2 links estrogen and Wnt signaling pathways by directly interacting with estrogen receptor-α (ERα) and β-catenin and thus promotes cell cycle progression. In breast cancer cells, independently of the SET domain that is involved in catalyzing H3K27me3, EZH2 transactivates the transcription of c-Myc and cyclinD1, thereby promoting cell proliferation. Neither EZH2 overexpression nor its knockdown changed the methylation status of H3K27 around c-Myc promoter. This activation event depends on the presence of ER, as activation of a reporter gene driven by c-Myc and cyclinD1 promoter was abolished by the addition of ER antagonist in ER-positive MCF-7 and T47-D cells. Furthermore, activation of reporter constructs was not observed in ER-negative MDA-MB-231 cells. This supports the importance of ERs in EZH2-mediated activation of target genes downstream of the estrogen signaling pathway. Since neither c-Myc nor cyclinD1 promoters contains estrogen responsive elements (EREs), ERα may interact via β-catenin, which can bind to multiple TCF/β-catenin binding sites. EZH2 integrates estrogen and Wnt pathways by forming a transcription complex with ERα, β-catenin and other co-activator proteins, such as SRC-1, and binding to the TCF/β-catenin binding sites on c-Myc and cyclinD1 promoters (). The in vivo interaction between ERα and EZH2 is estrogen-dependent and specific. In summary, based on the above experimental evidences, it is logical to propose that, when overexpressed during malignancy, EZH2 may function as a co-activator, being recruited to the estrogen-signaling pathway to enhance estrogen signaling and to promote proliferation. On the contrary, Hwang et al.Citation103 reported the identification of a novel binding partner for EZH2, the repressor of estrogen receptor activity (REA), and suggested that their interaction leads to the recruitment of EZH2 to ERE-containing target gene promoters and the repression of ER transcription. Based on the current evidence, it may be proposed that, depending on the cellular context, EZH2 functions as a multifaceted molecule acting either as a transcriptional repressor or activator in promoting breast tumorigenesis.
EZH2 in prostate carcinogenesis
Prostate cancer is the most common non-cutaneous cancer and major cause of cancer-related death among men in the United States. Despite considerable progress in cancer research, tumor heterogeneity and development of a hormone-refractory state continue to complicate diagnosis and treatment of prostate cancer. Highly sensitive and specific biomarkers are needed to distinguish indolent and aggressive tumors so that specific treatment strategies may be chosen based on the molecular profiling of a particular tumor. Also, molecular mechanisms regulating tumorigenesis and castration resistance need to be deciphered to develop novel therapeutic regimens targeting CRPC.
Like many other carcinomas, several genetic and epigenetic alterations have been associated with prostate tumorigenesis.Citation104 Earlier studies have shown only few candidate genes to be involved in prostate cancer, such as deletion or mutation in PTEN, p53, Nkx3.1, overexpression of oncogenes such as AR and GSTP1 promoter hypermethylation ().Citation105-Citation110 But after the advent of high throughput technologies and the completion of the human genome project, new molecular markers have been identified. Using cDNA microarray profiling, EZH2 was found to be overexpressed in invasive and hormone-refractory metastatic prostate cancers.Citation16 Recent reports suggest that EZH2 may promote prostate cancer progression by repressing tumor suppressor genes and developmental regulators and maintain a stem cell-like state by promoting a dedifferentiation program.Citation108,Citation111,Citation112 The molecular mechanism(s) reported to be responsible for the overexpression of EZH2 in prostate cancer include amplification of the EZH2 geneCitation113, the deletion of its negative regulator miR-101,Citation114 transcriptional regulation by MYCCitation115 and ETS gene family membersCitation129 as described in detail in .
EZH2 and androgen signaling
Androgens and the AR signaling pathway play a critical role in the normal development and functioning of the prostate, as well as in the proliferation and survival of prostate cancer cells.Citation113 Androgen deprivation causes involution of prostate tissue due to apoptosis in androgen-dependent epithelium. Similarly, androgen deprivation therapy (ADT) or castration kills prostate cancer cells that are dependent on androgens for their growth and survival. Nevertheless, after initial tumor remission, the disease relapses in a more lethal form called androgen-refractory or CRPC. The molecular mechanism by which androgens promote prostate tumorigenesis has not been comprehensively deciphered. Since EZH2 levels rise during advanced stages of prostate cancer, it is intriguing to investigate the role of androgens and AR signaling in regulating EZH2 expression. Bohrer et al.Citation114 have shown that EZH2 expression is repressed by androgens and that this repression requires a functional AR and is mediated through RB and p130-dependent pathways. This may provide a reason for elevated EZH2 expression in hormone-refractory and metastatic prostate cancer after ADT.
A recent study by Zhao et al.Citation115 highlights the previously overlooked role of AR as a global transcriptional repressor of several genes regulating cell differentiation and tumor suppression using a systems approach. The authors report that AR may play a dual role by inducing genes that promote prostatic differentiation while concomitantly repressing developmental regulators involved in non-prostatic pathways. Their study demonstrates that EZH2 collaborates with AR in this process. Since EZH2 is frequently overexpressed in metastatic prostate cancers, it may suppress AR-repressed genes in an androgen-deprived environment. Once recruited to the ARE motif-containing sequence, androgen receptor-dependent transcriptional silencing is mediated by EZH2 and subsequent chromatin remodeling. Their study supports the importance of adjuvant therapy targeting EZH2 along with hormone deprivation in advanced malignancies.
Cai et al.Citation116 reported that the synergy between AR and Kras signaling could elevate EZH2 levels, thereby promoting prostate tumorigenesis using a prostate tissue regeneration system. The crosstalk among EZH2, AR, ERG and HDACs upon androgen signaling was reported to promote prostate cancer progression.Citation117 ERG, together with HDACs and EZH2, modulate the transcriptional output of AR, thereby promoting tumorigenesis.
EZH2 and other endocrine related cancers
EZH2 has been implicated in a number of other endocrine-related cancers, such as pancreatic cancer, anaplastic thyroid carcinoma (ATC) and sporadic parathyroid adenoma. In pancreatic carcinogenesis, RAS upregulated EZH2 through MEK-ERK signaling, causing downregulation of tumor suppressors, including RUNX3.Citation118 In thyroid carcinoma, EZH2 overexpression is specific to ATC and directly controls ATC cells differentiation by causing transcriptional repression of the thyroid specific transcription factor paired-box gene 8 (PAX8).Citation119 Exome sequencing of tumors from patients with sporadic parathyroid adenomas revealed that the Y641N mutation in EZH2 was involved in molecular alterations causing sporadic parathyroid adenomas that cause primary hyperthyroidism.Citation120
EZH2 as Potential Cancer Therapeutic Target
Pharmacological and natural EZH2 inhibitors
Based on the current experimental evidences advocating the prominent role of EZH2 in tumor progression and metastatic spread, the search for novel and specific pharmacological inhibitors has yielded few interesting molecules. One of them, 3-dezaneplanocin-A (DZNep), is the most widely used EZH2 inhibitor and exhibits anti-tumor activity against a wide range of cancers, including breast, prostate, lung, brain and liver cancer cells.Citation121 Since EZH2-dependent histone methylation requires S-adenosyl-L-homocysteine (SAH) cofactor, it specifically targets SAH hydrolase and thus inhibits EZH2 function.Citation122 Despite its success as an anti-tumor agent targeting histone methylation and reactivating PRC2-repressed tumor suppressor genes in in vitro models, there are certain issues, such as its toxicity profile in animal models and selectivity, that need to be addressed. DZNep is a global methyltransferase inhibitor; therefore, EZH2-specific inhibitors are needed for a more effective response. Studies show that DZNep upregulated several anti-metastatic genes, thereby preventing cancer spreading, except in oral and ovarian cancers where genes upregulated by DZNep are also overexpressed in metastatic cells.Citation123 Therefore, targeting only EZH2 inhibition may not be an optimal universal treatment strategy for all tumor types. More recently, some EZH2 inhibitors have been developed that inhibit its enzymatic activity by directly binding to the enzyme and competing with the methyl group donor S-Adenosyl methionine (SAM) and are highly selective over EZH1 and other HMTs. EZH2 inhibitor EPZ005687, which has a greater selectivity and specificity, has been tested on lymphoma cells harboring tyrosine 641 or alanine 677 mutation in EZH2.Citation124 This compound reduced H3K27 trimethylation, which in turn resulted in apoptosis of mutant cancer cells with minimal effect on wild type cells. Wei Qi et al.Citation125 reported the small molecule inhibitor of EZH2, El1, which also acts in a SAM-competitive manner and reduce H3K27me3 levels.
Some dietary chemopreventive agents have been described as potent EZH2 inhibitors and can inhibit PRC2-mediated gene silencing. Dimri et al.Citation126 reported that dietary omega-3 (v-3) polyunsaturated fatty acids (PUFAs) can downregulate EZH2 expression and activity in breast cancer cells by inducing the ubiquitination and proteosomal degradation of EZH2. This PUFA-mediated inhibition resulted in the re-expression of known EZH2 targeted tumor suppressor genes such as E-cadherin and insulin-like growth factor binding protein, which ultimately decrease the invasiveness of breast cancer cells. Curcumin, a natural ingredient present in turmeric, also decrease breast cancer cell proliferation by modulating EZH2 levels and inducing G1 arrest in MDA-MB-435 breast cancer cells.Citation127 Mitogen-activated protein kinase (MAPK) pathway was reported to be involved in the downregulation of EZH2, contributing to the curcumin-induced anti-proliferative effect in breast cancer cells. A recent study demonstrated that (-)-epigallocatechin-3-gallate (EGCG), a major green tea polyphenol, independently or in combination with DZNep, reduces EZH2 and other PcG proteins such as EED, SUZ12, Mel18 and Bmi-1 levels in skin cancer cells.Citation128 Proteasome-dependent degradation of EZH2 and Bmi by EGCG and DZNep was the mechanisms proposed in the study. Interestingly, H3K27me3 levels were partially reduced in cells treated with EGCG or DZNep alone but there was a marked suppression in cells treated with both agents, suggesting that combined treatment could be more effective.
Conclusions
EZH2-mediated gene silencing acts in a highly interconnected and coordinated manner, epigenetically regulating a diverse cohort of genes involved in development, stem cell maintenance and cancer progression. In the past few years, significant efforts have been made to understand the oncogenic role of EZH2 in driving breast and prostate tumorigenesis. Interestingly, the interaction of EZH2 with AR or ER-mediated signaling pathways and the recently discovered activator role for EZH2 in CRPC and hormone-refractory breast cancer may provide attractive opportunities for therapeutic intervention to fight with the deadly forms of these diseases.
Future Perspectives
Targeting EZH2 inhibitors that directly suppress its activity by binding to its catalytic site and reducing global histone methylation levels may have detrimental side effects because EZH2 plays important roles in the normal functioning of the cell by maintaining the repressive state of many genes that function as developmental regulators and maintain stem cell pluripotency. Therefore, novel therapeutic strategies that focus on the regulatory circuit controlling EZH2 expression in a particular cancer cell-type or microenvironment can be beneficial. In this context, Chen et al.Citation41 demonstrated that inhibition of CDK1/2 mediated threonine 350 phosphorylation of EZH2 abolishes the global effect of EZH2 on gene silencing and proposed that CDK1/2 inhibitors may serve as alternative option to abrogate EZH2 activity in cancer cells. Furthermore, strategies should be pursued for designing specific PRC2 inhibitors targeting the EZH2 active site and/or surface for key subunit interactions. However, the diverse and complex nature of epigenetic regulatory network in cancer cells emphasizes the development of “combined therapeutic strategies” targeting multiple epigenetic pathways of tumor suppressor gene repression as well as their downstream effects in a particular type of cancer. Since EZH2, HDACs and DNMTs are parts of an interconnected epigenetic regulatory machinery, it becomes important to unravel the complete array of genes inactivated by their coordinated actions in cancer cells before proceeding to “combined treatment” strategies involving more than one epigenetic modifier.
| Abbreviations: | ||
| Enhancer of zeste homolog 1/2 | = | EZH1/2 |
| histone H3 lysine 27 trimethylation | = | H3K27me3 |
| mono-ubiquitination of Histone 2A at Lys 119 | = | H2AK119ub |
| polycomb repressive complex 2(PRC2) | = | PRC2 |
| embryonic stem cell | = | ESC |
| DNA methyltransferases | = | DNMTs |
| histone deacetylases | = | HDACs |
| castration resistant prostate cancer | = | CRPC |
| polycomb group protein | = | PcG |
| histone methyltransferases | = | HMTase |
| cysteine rich domain | = | CXC |
| su(var)3-9 enhancer of zeste trithorax domain | = | SET |
| non-coding RNA binding domain and a DNA binding domain | = | ncRBD |
| polycomb response elements | = | PREs |
| long non-coding RNAs | = | ncRNA |
| methyl CpG binding protein 2(MeCP2) | = | MeCP2 |
| methyl CpG binding protein 1 | = | MBD1 |
| methylated CpG island microarrays | = | MCAM |
| estrogen receptor | = | ER |
| androgen receptor(AR) | = | AR |
| progesterone receptor | = | PR |
| breast tumor initiating cells | = | BTICs |
| HIF response element | = | HRE |
| phosphoinositide-3-kinase/Akt | = | PI3K/Akt |
| estrogen response elements | = | EREs |
| repressor of estrogen receptor activity | = | REA |
| androgen deprivation therapy | = | ADT |
| anaplastic thyroid carcinoma | = | ADC |
| paired-box gene 8(PAX8) | = | PAX8 |
| 3-Dezaneplanocin-A(DZNep) | = | DZNep |
| S-adenosyl-L-homocysteine(SAH) | = | SAH |
| S-Adenosyl methionine | = | SAM |
| mitogen activated protein kinase | = | MAPK |
| epigallaocatechin-3-gallate | = | EGCG |
Acknowledgments
This work was partially supported by funds from the United States Public Health Service Grants RO1CA115491, RO1CA108512 and R21109424 to SG. We acknowledge Shyama Prasad Mukherjee (SPM) fellowship provided to GD by the Council of Scientific and Industrial Research (CSIR), India and Fulbright-Nehru Doctoral and Professional Research fellowship provided by United States – India Educational Foundation (USIEF) for her work in the United States.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011; 144:646 - 74; http://dx.doi.org/10.1016/j.cell.2011.02.013; PMID: 21376230
- You JS, Jones PA. Cancer genetics and epigenetics: two sides of the same coin?. Cancer Cell 2012; 22:9 - 20; http://dx.doi.org/10.1016/j.ccr.2012.06.008; PMID: 22789535
- Rodríguez-Paredes M, Esteller M. Cancer epigenetics reaches mainstream oncology. Nat Med 2011; 17:330 - 9; http://dx.doi.org/10.1038/nm.2305; PMID: 21386836
- Wilson BG, Roberts CW. SWI/SNF nucleosome remodellers and cancer. Nat Rev Cancer 2011; 11:481 - 92; http://dx.doi.org/10.1038/nrc3068; PMID: 21654818
- Jenuwein T, Laible G, Dorn R, Reuter G. SET domain proteins modulate chromatin domains in eu- and heterochromatin. Cell Mol Life Sci 1998; 54:80 - 93; http://dx.doi.org/10.1007/s000180050127; PMID: 9487389
- Tachibana M, Ueda J, Fukuda M, Takeda N, Ohta T, Iwanari H, et al. Histone methyltransferases G9a and GLP form heteromeric complexes and are both crucial for methylation of euchromatin at H3-K9. Genes Dev 2005; 19:815 - 26; http://dx.doi.org/10.1101/gad.1284005; PMID: 15774718
- Stewart MD, Li J, Wong J. Relationship between histone H3 lysine 9 methylation, transcription repression, and heterochromatin protein 1 recruitment. Mol Cell Biol 2005; 25:2525 - 38; http://dx.doi.org/10.1128/MCB.25.7.2525-2538.2005; PMID: 15767660
- Jenuwein T, Allis CD. Translating the histone code. Science 2001; 293:1074 - 80; http://dx.doi.org/10.1126/science.1063127; PMID: 11498575
- Cao R, Zhang Y. The functions of E(Z)/EZH2-mediated methylation of lysine 27 in histone H3. Curr Opin Genet Dev 2004; 14:155 - 64; http://dx.doi.org/10.1016/j.gde.2004.02.001; PMID: 15196462
- Schuettengruber B, Chourrout D, Vervoort M, Leblanc B, Cavalli G. Genome regulation by polycomb and trithorax proteins. Cell 2007; 128:735 - 45; http://dx.doi.org/10.1016/j.cell.2007.02.009; PMID: 17320510
- Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature 2006; 441:349 - 53; http://dx.doi.org/10.1038/nature04733; PMID: 16625203
- Sauvageau M, Sauvageau G. Polycomb group proteins: multi-faceted regulators of somatic stem cells and cancer. Cell Stem Cell 2010; 7:299 - 313; http://dx.doi.org/10.1016/j.stem.2010.08.002; PMID: 20804967
- Kondo Y, Shen L, Cheng AS, Ahmed S, Boumber Y, Charo C, et al. Gene silencing in cancer by histone H3 lysine 27 trimethylation independent of promoter DNA methylation. Nat Genet 2008; 40:741 - 50; http://dx.doi.org/10.1038/ng.159; PMID: 18488029
- Rush M, Appanah R, Lee S, Lam LL, Goyal P, Lorincz MC. Targeting of EZH2 to a defined genomic site is sufficient for recruitment of Dnmt3a but not de novo DNA methylation. Epigenetics 2009; 4:404 - 14; http://dx.doi.org/10.4161/epi.4.6.9392; PMID: 19717977
- Kleer CG, Cao Q, Varambally S, Shen R, Ota I, Tomlins SA, et al. EZH2 is a marker of aggressive breast cancer and promotes neoplastic transformation of breast epithelial cells. Proc Natl Acad Sci U S A 2003; 100:11606 - 11; http://dx.doi.org/10.1073/pnas.1933744100; PMID: 14500907
- Varambally S, Dhanasekaran SM, Zhou M, Barrette TR, Kumar-Sinha C, Sanda MG, et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature 2002; 419:624 - 9; http://dx.doi.org/10.1038/nature01075; PMID: 12374981
- Rau KM, Kang HY, Cha TL, Miller SA, Hung MC. The mechanisms and managements of hormone-therapy resistance in breast and prostate cancers. Endocr Relat Cancer 2005; 12:511 - 32; http://dx.doi.org/10.1677/erc.1.01026; PMID: 16172190
- Xu K, Wu ZJ, Groner AC, He HH, Cai C, Lis RT, et al. EZH2 oncogenic activity in castration-resistant prostate cancer cells is Polycomb-independent. Science 2012; 338:1465 - 9; http://dx.doi.org/10.1126/science.1227604; PMID: 23239736
- Cavalli G. Molecular biology. EZH2 goes solo. Science 2012; 338:1430 - 1; http://dx.doi.org/10.1126/science.1232332; PMID: 23239724
- Lee ST, Li Z, Wu Z, Aau M, Guan P, Karuturi RK, et al. Context-specific regulation of NF-κB target gene expression by EZH2 in breast cancers. Mol Cell 2011; 43:798 - 810; http://dx.doi.org/10.1016/j.molcel.2011.08.011; PMID: 21884980
- Shi B, Liang J, Yang X, Wang Y, Zhao Y, Wu H, et al. Integration of estrogen and Wnt signaling circuits by the polycomb group protein EZH2 in breast cancer cells. Mol Cell Biol 2007; 27:5105 - 19; http://dx.doi.org/10.1128/MCB.00162-07; PMID: 17502350
- Lewis EB. A gene complex controlling segmentation in Drosophila. Nature 1978; 276:565 - 70; http://dx.doi.org/10.1038/276565a0; PMID: 103000
- Yu M, Mazor T, Huang H, Huang HT, Kathrein KL, Woo AJ, et al. Direct recruitment of polycomb repressive complex 1 to chromatin by core binding transcription factors. Mol Cell 2012; 45:330 - 43; http://dx.doi.org/10.1016/j.molcel.2011.11.032; PMID: 22325351
- Levine SS, Weiss A, Erdjument-Bromage H, Shao Z, Tempst P, Kingston RE. The core of the polycomb repressive complex is compositionally and functionally conserved in flies and humans. Mol Cell Biol 2002; 22:6070 - 8; http://dx.doi.org/10.1128/MCB.22.17.6070-6078.2002; PMID: 12167701
- Wang L, Brown JL, Cao R, Zhang Y, Kassis JA, Jones RS. Hierarchical recruitment of polycomb group silencing complexes. Mol Cell 2004; 14:637 - 46; http://dx.doi.org/10.1016/j.molcel.2004.05.009; PMID: 15175158
- Yap KL, Li S, Muñoz-Cabello AM, Raguz S, Zeng L, Mujtaba S, et al. Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol Cell 2010; 38:662 - 74; http://dx.doi.org/10.1016/j.molcel.2010.03.021; PMID: 20541999
- Shao Z, Raible F, Mollaaghababa R, Guyon JR, Wu CT, Bender W, et al. Stabilization of chromatin structure by PRC1, a Polycomb complex. Cell 1999; 98:37 - 46; http://dx.doi.org/10.1016/S0092-8674(00)80604-2; PMID: 10412979
- Margueron R, Li G, Sarma K, Blais A, Zavadil J, Woodcock CL, et al. Ezh1 and Ezh2 maintain repressive chromatin through different mechanisms. Mol Cell 2008; 32:503 - 18; http://dx.doi.org/10.1016/j.molcel.2008.11.004; PMID: 19026781
- Kim H, Kang K, Kim J. AEBP2 as a potential targeting protein for Polycomb Repression Complex PRC2. Nucleic Acids Res 2009; 37:2940 - 50; http://dx.doi.org/10.1093/nar/gkp149; PMID: 19293275
- Nekrasov M, Klymenko T, Fraterman S, Papp B, Oktaba K, Köcher T, et al. Pcl-PRC2 is needed to generate high levels of H3-K27 trimethylation at Polycomb target genes. EMBO J 2007; 26:4078 - 88; http://dx.doi.org/10.1038/sj.emboj.7601837; PMID: 17762866
- Walker E, Chang WY, Hunkapiller J, Cagney G, Garcha K, Torchia J, et al. Polycomb-like 2 associates with PRC2 and regulates transcriptional networks during mouse embryonic stem cell self-renewal and differentiation. Cell Stem Cell 2010; 6:153 - 66; http://dx.doi.org/10.1016/j.stem.2009.12.014; PMID: 20144788
- Abel KJ, Brody LC, Valdes JM, Erdos MR, McKinley DR, Castilla LH, et al. Characterization of EZH1, a human homolog of Drosophila Enhancer of zeste near BRCA1. Genomics 1996; 37:161 - 71; http://dx.doi.org/10.1006/geno.1996.0537; PMID: 8921387
- Bracken AP, Pasini D, Capra M, Prosperini E, Colli E, Helin K. EZH2 is downstream of the pRB-E2F pathway, essential for proliferation and amplified in cancer. EMBO J 2003; 22:5323 - 35; http://dx.doi.org/10.1093/emboj/cdg542; PMID: 14532106
- Cardoso C, Mignon C, Hetet G, Grandchamps B, Fontes M, Colleaux L. The human EZH2 gene: genomic organisation and revised mapping in 7q35 within the critical region for malignant myeloid disorders. Eur J Hum Genet 2000; 8:174 - 80; http://dx.doi.org/10.1038/sj.ejhg.5200439; PMID: 10780782
- Margueron R, Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature 2011; 469:343 - 9; http://dx.doi.org/10.1038/nature09784; PMID: 21248841
- Chase A, Cross NC. Aberrations of EZH2 in cancer. Clin Cancer Res 2011; 17:2613 - 8; http://dx.doi.org/10.1158/1078-0432.CCR-10-2156; PMID: 21367748
- Morin RD, Johnson NA, Severson TM, Mungall AJ, An J, Goya R, et al. Somatic mutations altering EZH2 (Tyr641) in follicular and diffuse large B-cell lymphomas of germinal-center origin. Nat Genet 2010; 42:181 - 5; http://dx.doi.org/10.1038/ng.518; PMID: 20081860
- Ernst T, Chase AJ, Score J, Hidalgo-Curtis CE, Bryant C, Jones AV, et al. Inactivating mutations of the histone methyltransferase gene EZH2 in myeloid disorders. Nat Genet 2010; 42:722 - 6; http://dx.doi.org/10.1038/ng.621; PMID: 20601953
- Nikoloski G, Langemeijer SMC, Kuiper RP, Knops R, Massop M, Tönnissen ERLTM, et al. Somatic mutations of the histone methyltransferase gene EZH2 in myelodysplastic syndromes. Nat Genet 2010; 42:665 - 7; http://dx.doi.org/10.1038/ng.620; PMID: 20601954
- Cha TL, Zhou BP, Xia W, Wu Y, Yang CC, Chen CT, et al. Akt-mediated phosphorylation of EZH2 suppresses methylation of lysine 27 in histone H3. Science 2005; 310:306 - 10; http://dx.doi.org/10.1126/science.1118947; PMID: 16224021
- Chen S, Bohrer LR, Rai AN, Pan Y, Gan L, Zhou X, et al. Cyclin-dependent kinases regulate epigenetic gene silencing through phosphorylation of EZH2. Nat Cell Biol 2010; 12:1108 - 14; http://dx.doi.org/10.1038/ncb2116; PMID: 20935635
- Riising EM, Boggio R, Chiocca S, Helin K, Pasini D. The polycomb repressive complex 2 is a potential target of SUMO modifications. PLoS One 2008; 3:e2704; http://dx.doi.org/10.1371/journal.pone.0002704; PMID: 18628979
- Ku M, Koche RP, Rheinbay E, Mendenhall EM, Endoh M, Mikkelsen TS, et al. Genomewide analysis of PRC1 and PRC2 occupancy identifies two classes of bivalent domains. PLoS Genet 2008; 4:e1000242; http://dx.doi.org/10.1371/journal.pgen.1000242; PMID: 18974828
- Wilkinson FH, Park K, Atchison ML. Polycomb recruitment to DNA in vivo by the YY1 REPO domain. Proc Natl Acad Sci U S A 2006; 103:19296 - 301; http://dx.doi.org/10.1073/pnas.0603564103; PMID: 17158804
- Xi H, Yu Y, Fu Y, Foley J, Halees A, Weng Z. Analysis of overrepresented motifs in human core promoters reveals dual regulatory roles of YY1. Genome Res 2007; 17:798 - 806; http://dx.doi.org/10.1101/gr.5754707; PMID: 17567998
- Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 2007; 129:1311 - 23; http://dx.doi.org/10.1016/j.cell.2007.05.022; PMID: 17604720
- Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science 2010; 329:689 - 93; http://dx.doi.org/10.1126/science.1192002; PMID: 20616235
- Plath K, Fang J, Mlynarczyk-Evans SK, Cao R, Worringer KA, Wang H, et al. Role of histone H3 lysine 27 methylation in X inactivation. Science 2003; 300:131 - 5; http://dx.doi.org/10.1126/science.1084274; PMID: 12649488
- Maenner S, Blaud M, Fouillen L, Savoye A, Marchand V, Dubois A, et al. 2-D structure of the A region of Xist RNA and its implication for PRC2 association. PLoS Biol 2010; 8:e1000276; http://dx.doi.org/10.1371/journal.pbio.1000276; PMID: 20052282
- Kohlmaier A, Savarese F, Lachner M, Martens J, Jenuwein T, Wutz A. A chromosomal memory triggered by Xist regulates histone methylation in X inactivation. PLoS Biol 2004; 2:E171; http://dx.doi.org/10.1371/journal.pbio.0020171; PMID: 15252442
- Pandey RR, Mondal T, Mohammad F, Enroth S, Redrup L, Komorowski J, et al. Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol Cell 2008; 32:232 - 46; http://dx.doi.org/10.1016/j.molcel.2008.08.022; PMID: 18951091
- Eskeland R, Leeb M, Grimes GR, Kress C, Boyle S, Sproul D, et al. Ring1B compacts chromatin structure and represses gene expression independent of histone ubiquitination. Mol Cell 2010; 38:452 - 64; http://dx.doi.org/10.1016/j.molcel.2010.02.032; PMID: 20471950
- Schoeftner S, Sengupta AK, Kubicek S, Mechtler K, Spahn L, Koseki H, et al. Recruitment of PRC1 function at the initiation of X inactivation independent of PRC2 and silencing. EMBO J 2006; 25:3110 - 22; http://dx.doi.org/10.1038/sj.emboj.7601187; PMID: 16763550
- Ko S, Ahn J, Song CS, Kim S, Knapczyk-Stwora K, Chatterjee B. Lysine methylation and functional modulation of androgen receptor by Set9 methyltransferase. Mol Endocrinol 2011; 25:433 - 44; http://dx.doi.org/10.1210/me.2010-0482; PMID: 21273441
- Gaughan L, Stockley J, Wang N, McCracken SRC, Treumann A, Armstrong K, et al. Regulation of the androgen receptor by SET9-mediated methylation. Nucleic Acids Res 2011; 39:1266 - 79; http://dx.doi.org/10.1093/nar/gkq861; PMID: 20959290
- Nan X, Ng H-H, Johnson CA, Laherty CD, Turner BM, Eisenman RN, et al. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature 1998; 393:386 - 9; http://dx.doi.org/10.1038/30764; PMID: 9620804
- Jones PL, Veenstra GJ, Wade PA, Vermaak D, Kass SU, Landsberger N, et al. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat Genet 1998; 19:187 - 91; http://dx.doi.org/10.1038/561; PMID: 9620779
- Simon JA, Lange CA. Roles of the EZH2 histone methyltransferase in cancer epigenetics. Mutat Res 2008; 647:21 - 9; http://dx.doi.org/10.1016/j.mrfmmm.2008.07.010; PMID: 18723033
- Viré E, Brenner C, Deplus R, Blanchon L, Fraga M, Didelot C, et al. The Polycomb group protein EZH2 directly controls DNA methylation. Nature 2006; 439:871 - 4; http://dx.doi.org/10.1038/nature04431; PMID: 16357870
- McGarvey KM, Greene E, Fahrner JA, Jenuwein T, Baylin SB. DNA methylation and complete transcriptional silencing of cancer genes persist after depletion of EZH2. Cancer Res 2007; 67:5097 - 102; http://dx.doi.org/10.1158/0008-5472.CAN-06-2029; PMID: 17545586
- Ohm JE, McGarvey KM, Yu X, Cheng L, Schuebel KE, Cope L, et al. A stem cell-like chromatin pattern may predispose tumor suppressor genes to DNA hypermethylation and heritable silencing. Nat Genet 2007; 39:237 - 42; http://dx.doi.org/10.1038/ng1972; PMID: 17211412
- Schlesinger Y, Straussman R, Keshet I, Farkash S, Hecht M, Zimmerman J, et al. Polycomb-mediated methylation on Lys27 of histone H3 pre-marks genes for de novo methylation in cancer. Nat Genet 2007; 39:232 - 6; http://dx.doi.org/10.1038/ng1950; PMID: 17200670
- Widschwendter M, Fiegl H, Egle D, Mueller-Holzner E, Spizzo G, Marth C, et al. Epigenetic stem cell signature in cancer. Nat Genet 2007; 39:157 - 8; http://dx.doi.org/10.1038/ng1941; PMID: 17200673
- Barski A, Cuddapah S, Cui K, Roh T-Y, Schones DE, Wang Z, et al. High-resolution profiling of histone methylations in the human genome. Cell 2007; 129:823 - 37; http://dx.doi.org/10.1016/j.cell.2007.05.009; PMID: 17512414
- Bell O, Wirbelauer C, Hild M, Scharf AND, Schwaiger M, MacAlpine DM, et al. Localized H3K36 methylation states define histone H4K16 acetylation during transcriptional elongation in Drosophila. EMBO J 2007; 26:4974 - 84; http://dx.doi.org/10.1038/sj.emboj.7601926; PMID: 18007591
- Barrand S, Andersen IS, Collas P. Promoter-exon relationship of H3 lysine 9, 27, 36 and 79 methylation on pluripotency-associated genes. Biochem Biophys Res Commun 2010; 401:611 - 7; http://dx.doi.org/10.1016/j.bbrc.2010.09.116; PMID: 20920475
- Schmitges FW, Prusty AB, Faty M, Stützer A, Lingaraju GM, Aiwazian J, et al. Histone methylation by PRC2 is inhibited by active chromatin marks. Mol Cell 2011; 42:330 - 41; http://dx.doi.org/10.1016/j.molcel.2011.03.025; PMID: 21549310
- Yuan W, Xu M, Huang C, Liu N, Chen S, Zhu B. H3K36 methylation antagonizes PRC2-mediated H3K27 methylation. J Biol Chem 2011; 286:7983 - 9; http://dx.doi.org/10.1074/jbc.M110.194027; PMID: 21239496
- Tie F, Banerjee R, Stratton CA, Prasad-Sinha J, Stepanik V, Zlobin A, et al. CBP-mediated acetylation of histone H3 lysine 27 antagonizes Drosophila Polycomb silencing. Development 2009; 136:3131 - 41; http://dx.doi.org/10.1242/dev.037127; PMID: 19700617
- Pasini D, Malatesta M, Jung HR, Walfridsson J, Willer A, Olsson L, et al. Characterization of an antagonistic switch between histone H3 lysine 27 methylation and acetylation in the transcriptional regulation of Polycomb group target genes. Nucleic Acids Res 2010; 38:4958 - 69; http://dx.doi.org/10.1093/nar/gkq244; PMID: 20385584
- Schwartz YB, Kahn TG, Stenberg P, Ohno K, Bourgon R, Pirrotta V. Alternative epigenetic chromatin states of polycomb target genes. PLoS Genet 2010; 6:e1000805; http://dx.doi.org/10.1371/journal.pgen.1000805; PMID: 20062800
- Lau PNI, Cheung P. Histone code pathway involving H3 S28 phosphorylation and K27 acetylation activates transcription and antagonizes polycomb silencing. Proc Natl Acad Sci U S A 2011; 108:2801 - 6; http://dx.doi.org/10.1073/pnas.1012798108; PMID: 21282660
- Gehani SS, Agrawal-Singh S, Dietrich N, Christophersen NS, Helin K, Hansen K. Polycomb group protein displacement and gene activation through MSK-dependent H3K27me3S28 phosphorylation. Mol Cell 2010; 39:886 - 900; http://dx.doi.org/10.1016/j.molcel.2010.08.020; PMID: 20864036
- O’Meara MM, Simon JA. Inner workings and regulatory inputs that control Polycomb repressive complex 2. Chromosoma 2012; 121:221 - 34; http://dx.doi.org/10.1007/s00412-012-0361-1; PMID: 22349693
- DeSantis C, Siegel R, Bandi P, Jemal A. Breast cancer statistics, 2011. CA Cancer J Clin 2011; 61:409 - 18; http://dx.doi.org/10.3322/caac.20134; PMID: 21969133
- Ross JS, Fletcher JA. The HER-2/neu Oncogene in Breast Cancer: Prognostic Factor, Predictive Factor, and Target for Therapy. Oncologist 1998; 3:237 - 52; PMID: 10388110
- Hayes DF, Trock B, Harris AL. Assessing the clinical impact of prognostic factors: when is “statistically significant” clinically useful?. Breast Cancer Res Treat 1998; 52:305 - 19; http://dx.doi.org/10.1023/A:1006197805041; PMID: 10066089
- Hayes DF. Do we need prognostic factors in nodal-negative breast cancer? Arbiter. Eur J Cancer 2000; 36:302 - 6; http://dx.doi.org/10.1016/S0959-8049(99)00303-2; PMID: 10708930
- Collett K, Eide GE, Arnes J, Stefansson IM, Eide J, Braaten A, et al. Expression of enhancer of zeste homologue 2 is significantly associated with increased tumor cell proliferation and is a marker of aggressive breast cancer. Clin Cancer Res 2006; 12:1168 - 74; http://dx.doi.org/10.1158/1078-0432.CCR-05-1533; PMID: 16489070
- Bachmann IM, Halvorsen OJ, Collett K, Stefansson IM, Straume O, Haukaas SA, et al. EZH2 expression is associated with high proliferation rate and aggressive tumor subgroups in cutaneous melanoma and cancers of the endometrium, prostate, and breast. J Clin Oncol 2006; 24:268 - 73; http://dx.doi.org/10.1200/JCO.2005.01.5180; PMID: 16330673
- Gonzalez ME, Li X, Toy K, DuPrie M, Ventura AC, Banerjee M, et al. Downregulation of EZH2 decreases growth of estrogen receptor-negative invasive breast carcinoma and requires BRCA1. Oncogene 2009; 28:843 - 53; http://dx.doi.org/10.1038/onc.2008.433; PMID: 19079346
- Du J, Li L, Ou Z, Kong C, Zhang Y, Dong Z, et al. FOXC1, a target of polycomb, inhibits metastasis of breast cancer cells. Breast Cancer Res Treat 2012; 131:65 - 73; http://dx.doi.org/10.1007/s10549-011-1396-3; PMID: 21465172
- Zeidler M, Varambally S, Cao Q, Chinnaiyan AM, Ferguson DO, Merajver SD, et al. The Polycomb group protein EZH2 impairs DNA repair in breast epithelial cells. Neoplasia 2005; 7:1011 - 9; http://dx.doi.org/10.1593/neo.05472; PMID: 16331887
- Yang X, Karuturi RK, Sun F, Aau M, Yu K, Shao R, et al. CDKN1C (p57) is a direct target of EZH2 and suppressed by multiple epigenetic mechanisms in breast cancer cells. PLoS One 2009; 4:e5011; http://dx.doi.org/10.1371/journal.pone.0005011; PMID: 19340297
- Fujii S, Ito K, Ito Y, Ochiai A. Enhancer of zeste homologue 2 (EZH2) down-regulates RUNX3 by increasing histone H3 methylation. J Biol Chem 2008; 283:17324 - 32; http://dx.doi.org/10.1074/jbc.M800224200; PMID: 18430739
- Ren G, Baritaki S, Marathe H, Feng J, Park S, Beach S, et al. Polycomb protein EZH2 regulates tumor invasion via the transcriptional repression of the metastasis suppressor RKIP in breast and prostate cancer. Cancer Res 2012; 72:3091 - 104; http://dx.doi.org/10.1158/0008-5472.CAN-11-3546; PMID: 22505648
- Truax AD, Thakkar M, Greer SF. Dysregulated recruitment of the histone methyltransferase EZH2 to the class II transactivator (CIITA) promoter IV in breast cancer cells. PLoS One 2012; 7:e36013; http://dx.doi.org/10.1371/journal.pone.0036013; PMID: 22563434
- Taniguchi H, Jacinto FV, Villanueva A, Fernandez AF, Yamamoto H, Carmona FJ, et al. Silencing of Kruppel-like factor 2 by the histone methyltransferase EZH2 in human cancer. Oncogene 2012; 31:1988 - 94; http://dx.doi.org/10.1038/onc.2011.387; PMID: 21892211
- Gonzalez ME, DuPrie ML, Krueger H, Merajver SD, Ventura AC, Toy KA, et al. Histone methyltransferase EZH2 induces Akt-dependent genomic instability and BRCA1 inhibition in breast cancer. Cancer Res 2011; 71:2360 - 70; http://dx.doi.org/10.1158/0008-5472.CAN-10-1933; PMID: 21406404
- Dyson N. The regulation of E2F by pRB-family proteins. Genes Dev 1998; 12:2245 - 62; http://dx.doi.org/10.1101/gad.12.15.2245; PMID: 9694791
- Helin K. Regulation of cell proliferation by the E2F transcription factors. Curr Opin Genet Dev 1998; 8:28 - 35; http://dx.doi.org/10.1016/S0959-437X(98)80058-0; PMID: 9529602
- Trimarchi JM, Lees JA. Sibling rivalry in the E2F family. Nat Rev Mol Cell Biol 2002; 3:11 - 20; http://dx.doi.org/10.1038/nrm714; PMID: 11823794
- Bracken AP, Pasini D, Capra M, Prosperini E, Colli E, Helin K. EZH2 is downstream of the pRB-E2F pathway, essential for proliferation and amplified in cancer. EMBO J 2003; 22:5323 - 35; http://dx.doi.org/10.1093/emboj/cdg542; PMID: 14532106
- Fujii S, Tokita K, Wada N, Ito K, Yamauchi C, Ito Y, et al. MEK-ERK pathway regulates EZH2 overexpression in association with aggressive breast cancer subtypes. Oncogene 2011; 30:4118 - 28; http://dx.doi.org/10.1038/onc.2011.118; PMID: 21499305
- Chang CJ, Yang JY, Xia W, Chen CT, Xie X, Chao CH, et al. EZH2 promotes expansion of breast tumor initiating cells through activation of RAF1-β-catenin signaling. Cancer Cell 2011; 19:86 - 100; http://dx.doi.org/10.1016/j.ccr.2010.10.035; PMID: 21215703
- Venkitaraman AR. Cancer susceptibility and the functions of BRCA1 and BRCA2. Cell 2002; 108:171 - 82; http://dx.doi.org/10.1016/S0092-8674(02)00615-3; PMID: 11832208
- Narod SA, Foulkes WD. BRCA1 and BRCA2: 1994 and beyond. Nat Rev Cancer 2004; 4:665 - 76; http://dx.doi.org/10.1038/nrc1431; PMID: 15343273
- Wilson CA, Ramos L, Villaseñor MR, Anders KH, Press MF, Clarke K, et al. Localization of human BRCA1 and its loss in high-grade, non-inherited breast carcinomas. Nat Genet 1999; 21:236 - 40; http://dx.doi.org/10.1038/6029; PMID: 9988281
- Turner NC, Reis-Filho JS, Russell AM, Springall RJ, Ryder K, Steele D, et al. BRCA1 dysfunction in sporadic basal-like breast cancer. Oncogene 2007; 26:2126 - 32; http://dx.doi.org/10.1038/sj.onc.1210014; PMID: 17016441
- Foulkes WD, Stefansson IM, Chappuis PO, Bégin LR, Goffin JR, Wong N, et al. Germline BRCA1 mutations and a basal epithelial phenotype in breast cancer. J Natl Cancer Inst 2003; 95:1482 - 5; http://dx.doi.org/10.1093/jnci/djg050; PMID: 14519755
- Lakhani SR, Reis-Filho JS, Fulford L, Penault-Llorca F, van der Vijver M, Parry S, et al, Breast Cancer Linkage Consortium. Prediction of BRCA1 status in patients with breast cancer using estrogen receptor and basal phenotype. Clin Cancer Res 2005; 11:5175 - 80; http://dx.doi.org/10.1158/1078-0432.CCR-04-2424; PMID: 16033833
- Hervouet E, Cartron P-F, Jouvenot M, Delage-Mourroux R. Epigenetic regulation of estrogen signaling in breast cancer. Epigenetics 2013; 8:237 - 45; http://dx.doi.org/10.4161/epi.23790; PMID: 23364277
- Hwang C, Giri VN, Wilkinson JC, Wright CW, Wilkinson AS, Cooney KA, et al. EZH2 regulates the transcription of estrogen-responsive genes through association with REA, an estrogen receptor corepressor. Breast Cancer Res Treat 2008; 107:235 - 42; http://dx.doi.org/10.1007/s10549-007-9542-7; PMID: 17453341
- Schulz WA, Hoffmann MJ. Epigenetic mechanisms in the biology of prostate cancer. Semin Cancer Biol 2009; 19:172 - 80; http://dx.doi.org/10.1016/j.semcancer.2009.02.006; PMID: 19429481
- Chen H, Tu SW, Hsieh JT. Down-regulation of human DAB2IP gene expression mediated by polycomb Ezh2 complex and histone deacetylase in prostate cancer. J Biol Chem 2005; 280:22437 - 44; http://dx.doi.org/10.1074/jbc.M501379200; PMID: 15817459
- Beke L, Nuytten M, Van Eynde A, Beullens M, Bollen M. The gene encoding the prostatic tumor suppressor PSP94 is a target for repression by the Polycomb group protein EZH2. Oncogene 2007; 26:4590 - 5; http://dx.doi.org/10.1038/sj.onc.1210248; PMID: 17237810
- Yu J, Cao Q, Yu J, Wu L, Dallol A, Li J, et al. The neuronal repellent SLIT2 is a target for repression by EZH2 in prostate cancer. Oncogene 2010; 29:5370 - 80; http://dx.doi.org/10.1038/onc.2010.269; PMID: 20622896
- Cao Q, Yu J, Dhanasekaran SM, Kim JH, Mani RS, Tomlins SA, et al. Repression of E-cadherin by the polycomb group protein EZH2 in cancer. Oncogene 2008; 27:7274 - 84; http://dx.doi.org/10.1038/onc.2008.333; PMID: 18806826
- Yu J, Cao Q, Mehra R, Laxman B, Yu J, Tomlins SA, et al. Integrative genomics analysis reveals silencing of beta-adrenergic signaling by polycomb in prostate cancer. Cancer Cell 2007; 12:419 - 31; http://dx.doi.org/10.1016/j.ccr.2007.10.016; PMID: 17996646
- Shin YJ, Kim JH. The role of EZH2 in the regulation of the activity of matrix metalloproteinases in prostate cancer cells. PLoS One 2012; 7:e30393; http://dx.doi.org/10.1371/journal.pone.0030393; PMID: 22272343
- Yu J, Yu J, Rhodes DR, Tomlins SA, Cao X, Chen G, et al. A polycomb repression signature in metastatic prostate cancer predicts cancer outcome. Cancer Res 2007; 67:10657 - 63; http://dx.doi.org/10.1158/0008-5472.CAN-07-2498; PMID: 18006806
- Yu J, Yu J, Mani RS, Cao Q, Brenner CJ, Cao X, et al. An integrated network of androgen receptor, polycomb, and TMPRSS2-ERG gene fusions in prostate cancer progression. Cancer Cell 2010; 17:443 - 54; http://dx.doi.org/10.1016/j.ccr.2010.03.018; PMID: 20478527
- Shen MM, Abate-Shen C. Molecular genetics of prostate cancer: new prospects for old challenges. Genes Dev 2010; 24:1967 - 2000; http://dx.doi.org/10.1101/gad.1965810; PMID: 20844012
- Bohrer LR, Chen S, Hallstrom TC, Huang H. Androgens suppress EZH2 expression via retinoblastoma (RB) and p130-dependent pathways: a potential mechanism of androgen-refractory progression of prostate cancer. Endocrinology 2010; 151:5136 - 45; http://dx.doi.org/10.1210/en.2010-0436; PMID: 20881251
- Zhao JC, Yu J, Runkle C, Wu L, Hu M, Wu D, et al. Cooperation between Polycomb and androgen receptor during oncogenic transformation. Genome Res 2012; 22:322 - 31; http://dx.doi.org/10.1101/gr.131508.111; PMID: 22179855
- Cai H, Memarzadeh S, Stoyanova T, Beharry Z, Kraft AS, Witte ON. Collaboration of Kras and androgen receptor signaling stimulates EZH2 expression and tumor-propagating cells in prostate cancer. Cancer Res 2012; 72:4672 - 81; http://dx.doi.org/10.1158/0008-5472.CAN-12-0228; PMID: 22805308
- Chng KR, Chang CW, Tan SK, Yang C, Hong SZ, Sng NY, et al. A transcriptional repressor co-regulatory network governing androgen response in prostate cancers. EMBO J 2012; 31:2810 - 23; http://dx.doi.org/10.1038/emboj.2012.112; PMID: 22531786
- Fujii S, Fukamachi K, Tsuda H, Ito K, Ito Y, Ochiai A. RAS oncogenic signal upregulates EZH2 in pancreatic cancer. Biochem Biophys Res Commun 2012; 417:1074 - 9; http://dx.doi.org/10.1016/j.bbrc.2011.12.099; PMID: 22222375
- Borbone E, Troncone G, Ferraro A, Jasencakova Z, Stojic L, Esposito F, et al. Enhancer of zeste homolog 2 overexpression has a role in the development of anaplastic thyroid carcinomas. J Clin Endocrinol Metab 2011; 96:1029 - 38; http://dx.doi.org/10.1210/jc.2010-1784; PMID: 21289264
- Cromer MK, Starker LF, Choi M, Udelsman R, Nelson-Williams C, Lifton RP, et al. Identification of somatic mutations in parathyroid tumors using whole-exome sequencing. J Clin Endocrinol Metab 2012; 97:E1774 - 81; http://dx.doi.org/10.1210/jc.2012-1743; PMID: 22740705
- Tan J, Yang X, Zhuang L, Jiang X, Chen W, Lee PL, et al. Pharmacologic disruption of Polycomb-repressive complex 2-mediated gene repression selectively induces apoptosis in cancer cells. Genes Dev 2007; 21:1050 - 63; http://dx.doi.org/10.1101/gad.1524107; PMID: 17437993
- Miranda TB, Cortez CC, Yoo CB, Liang G, Abe M, Kelly TK, et al. DZNep is a global histone methylation inhibitor that reactivates developmental genes not silenced by DNA methylation. Mol Cancer Ther 2009; 8:1579 - 88; http://dx.doi.org/10.1158/1535-7163.MCT-09-0013; PMID: 19509260
- Crea F, Fornaro L, Bocci G, Sun L, Farrar WL, Falcone A, et al. EZH2 inhibition: targeting the crossroad of tumor invasion and angiogenesis. Cancer Metastasis Rev 2012; 31:753 - 61; http://dx.doi.org/10.1007/s10555-012-9387-3; PMID: 22711031
- Knutson SK, Wigle TJ, Warholic NM, Sneeringer CJ, Allain CJ, Klaus CR, et al. A selective inhibitor of EZH2 blocks H3K27 methylation and kills mutant lymphoma cells. Nat Chem Biol 2012; 8:890 - 6; PMID: 23023262
- Qi W, Chan H, Teng L, Li L, Chuai S, Zhang R, et al. Selective inhibition of Ezh2 by a small molecule inhibitor blocks tumor cells proliferation. Proc Natl Acad Sci U S A 2012; 109:21360 - 5; http://dx.doi.org/10.1073/pnas.1210371110; PMID: 23236167
- Dimri M, Bommi PV, Sahasrabuddhe AA, Khandekar JD, Dimri GP. Dietary omega-3 polyunsaturated fatty acids suppress expression of EZH2 in breast cancer cells. Carcinogenesis 2010; 31:489 - 95; http://dx.doi.org/10.1093/carcin/bgp305; PMID: 19969553
- Hua W-F, Fu Y-S, Liao Y-J, Xia W-J, Chen Y-C, Zeng Y-X, et al. Curcumin induces down-regulation of EZH2 expression through the MAPK pathway in MDA-MB-435 human breast cancer cells. Eur J Pharmacol 2010; 637:16 - 21; http://dx.doi.org/10.1016/j.ejphar.2010.03.051; PMID: 20385124
- Nandakumar V, Vaid M, Katiyar SK. (-)-Epigallocatechin-3-gallate reactivates silenced tumor suppressor genes, Cip1/p21 and p16INK4a, by reducing DNA methylation and increasing histones acetylation in human skin cancer cells. Carcinogenesis 2011; 32:537 - 44; http://dx.doi.org/10.1093/carcin/bgq285; PMID: 21209038
- Kunderfranco P, Mello-Grand M, Cangemi R, Mensah A, Albertini V, et al. ETS transcription factors control transcription of EZH2 and epigenetic silencing of the tumor suppressor gene Nkx3.1 in prostate cancer.. PloS one 2010; 5:e10547