Abstract
Schizophrenia (SCZ) is a devastating psychiatric disorder with a median lifetime prevalence rate of 0.7–0.8%. Elevated plasma total homocysteine has been suggested as a risk factor for SCZ, and various biological effects of hyperhomocysteinemia have been proposed to be relevant to the pathophysiology of SCZ. As increased attention is paid to aberrant DNA methylation in SCZ, homocysteine is attracting additional interest as a potential key substance. Homocysteine is formed in the methionine cycle, which is involved in one-carbon methyl group-transfer metabolism, and it acts as a methyl donor when it is converted to S-adenosyl-methionine. To date, no studies have examined the relationship between homocysteine and genome-wide DNA methylation in SCZ. We examined the relationship between plasma total homocysteine and DNA methylation patterns in the peripheral leukocytes of patients with SCZ (n = 42) using a quantitative high-resolution DNA methylation array (485,764 CpG sites). Significant homocysteine-related changes in DNA methylation were observed at 1,338 CpG sites that were located across whole gene regions, including promoters, gene bodies and 3′-untranslated regions. Of the 1,338 sites, 758 sites (56.6%) were located in the CpG islands (CGIs) and in the regions flanking CGIs (CGI: 15.8%; CGI shore: 28.2%; CGI shelf: 12.6%), and positive correlations between plasma total homocysteine and DNA methylation were observed predominantly at CpG sites in the CGIs. Our results suggest that homocysteine might play a role in the pathogenesis of SCZ via a molecular mechanism that involves alterations to DNA methylation.
Introduction
Schizophrenia (SCZ) is a devastating psychiatric disorder with a median lifetime prevalence rate of 0.7–0.8%.Citation1 Elevated plasma total homocysteine has been suggested as a risk factor for SCZ,Citation2,Citation3 and hyperhomocysteinemia has been proposed to contribute to the pathophysiology of SCZ via various biological effects, such as a partial antagonist of the glutamate site of the N-methyl-D-aspartate receptor,Citation4 the interferer of oxygen delivery by damaging placental vasculature,Citation2 DNA damage and cell cytotoxicity,Citation5 neuronal apoptosisCitation6 and mitochondrial nitric oxide accumulation.Citation7
Recently, accumulating evidence has shown that DNA methylation is also implicated in SCZ.Citation8-Citation27 As more attention is paid to DNA methylation, homocysteine has been recognized as a potentially key substance. Homocysteine is formed during the methionine cycle, is involved in one-carbon methyl group-transfer metabolism and acts as a methyl donor when it is converted to S-adenosyl-methionine. Several studies have reported an association between hyperhomocysteinemia and aberrant DNA methylation in several diseases, including atherosclerosis, osteoporosis, uremia and alcoholism.Citation28-Citation31 Furthermore, Fryer and colleagues reported a significant correlation between cord blood-plasma total homocysteine and DNA methylation at numerous CpG sites.Citation32 These studies led us to hypothesize that hyperhomocysteinemia in SCZ might have an impact on the DNA methylation levels in specific genes. However, to date, there are no reports that examine the relationship between homocysteine and genome-wide DNA methylation in SCZ.
To gain further insight into the pathogenic mechanisms that underlie hyperhomocysteinemia in SCZ, we examined the relationship between plasma total homocysteine and DNA methylation patterns in the peripheral leukocytes of patients with SCZ by using a quantitative high-resolution DNA methylation array.
Results
Differences in plasma total homocysteine between patients with SCZ and controls.
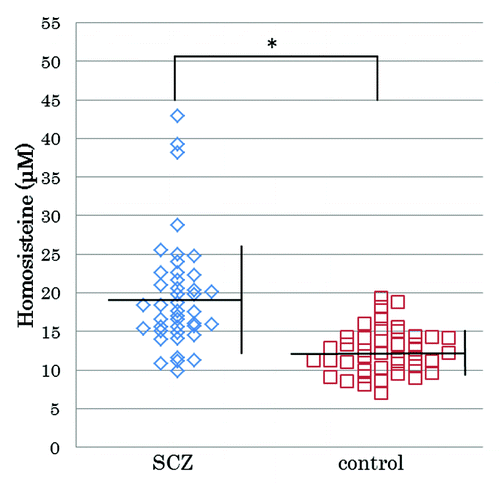
The mean plasma total homocysteine level in patients with SCZ (n = 42) was 19.5 ± 7.2 nmol/mL (mean ± SD), and the level in the control subjects (n = 42) was 12.4 ± 2.9 nmol/mL (mean ± SD). The plasma total homocysteine levels of the patient group were significantly higher than those of the control group (p < 0.0001), as shown in .
Figure 1. Plasma total homocysteine levels of patients with schizophrenia and controls. Blue dots represent plasma total homocysteine levels of patients with SCZ. Red dots represent plasma total homocysteine levels of controls. The mean plasma total homocysteine level in patients with SCZ (n = 42) was 19.5 ± 7.2 nmol/mL (mean ± SD), and the level in the control subjects (n = 42) was 12.4 ± 2.9 nmol/mL (mean ± SD). The plasma total homocysteine levels of the patient group were significant higher than those of the control group (Mann–Whitney U test, p < 0.0001)
Relationship between plasma total homocysteine and genome-wide DNA methylation patterns in patients with SCZ.
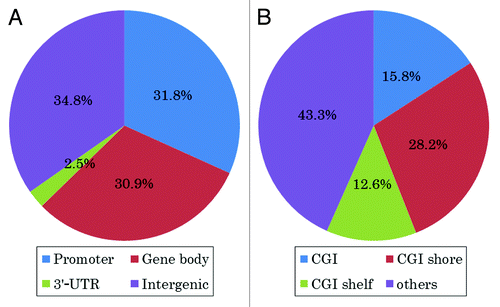
Of the 164,657 CpG sites analyzed, significant plasma total homocysteine-related changes in DNA methylation were observed at 1,338 sites (p < 0.01). The top 10-ranking CpG sites significantly associated with plasma total homocysteine are shown in , and the top 100-ranking CpG sites are shown in Table S1. Examples include two CpG sites in the SLC18A2 and the GNAL genes (). Both of these genes have been implicated in SCZ.Citation33-Citation36 When these 1,338 CpG sites were classified into four different categories according to their location in the genes [promoter, gene body, 3′- untranslated regions (UTRs) and intergenic region], 425 sites (31.8%) were located in the promoter regions, 414 sites (30.9%) in gene bodies and 34 sites (2.5%) in 3′-UTRs (). When these 1,338 CpG sites were classified into four categories according to the CpG content in the genes [CpG island (CGI), CGI shore, CGI shelf and others], 212 sites (15.8%) were located in the CGIs, 377 sites (28.2%) in CGI shores and 169 sites (12.6%) in CGI shelves (; Table S2). Of the significant 212 CpG sites in the CGIs, 74 sites (34.9%) were located in the promoter regions.
Table 1. The top 10-ranking of CpG sites significant associated with plasma homocysteine
Figure 2. Two CpG sites in the SLC18A2 (cg00498305) and GNAL (cg12327405) genes, which have been implicated in SCZ. A significant positive correlation of plasma total homocysteine with DNA methylation was observed at cg00498305 located in the CGI shore in the promoter region of the SLC18A2 gene (p = 1.67E-03). A significant negative correlation of plasma total homocysteine with DNA methylation was observed at cg12327405 in the CGI in the promoter region of the GNAL gene (p = 2.85E-04). [X-axis: plasma total homocysteine (nmol/mL); Y-axis: DNA methylation (β)]
![Figure 2. Two CpG sites in the SLC18A2 (cg00498305) and GNAL (cg12327405) genes, which have been implicated in SCZ. A significant positive correlation of plasma total homocysteine with DNA methylation was observed at cg00498305 located in the CGI shore in the promoter region of the SLC18A2 gene (p = 1.67E-03). A significant negative correlation of plasma total homocysteine with DNA methylation was observed at cg12327405 in the CGI in the promoter region of the GNAL gene (p = 2.85E-04). [X-axis: plasma total homocysteine (nmol/mL); Y-axis: DNA methylation (β)]](/cms/asset/327f8e60-5733-4e69-8536-7938786ae1fb/kepi_a_10924621_f0002.gif)
Figure 3. Percentages of 1,338 CpG sites at which plasma total homocysteine and DNA methylation were significantly correlated. (A) Of the 1,338 CpG sites, 425 (31.8%) were located in promoter regions, 414 (30.9%) were located in gene bodies and 34 (2.5%) were located in 3′−UTRs. (B) Of the 1,338 CpG sites, 212 (15.8%) were located in CGIs, 377 (28.2%) were located in CGI shores and 169 (12.6%) were located in CGI shelves.
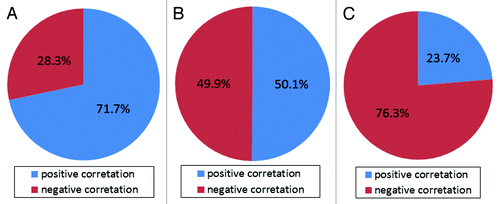
Of the 1,338 significant CpG sites, positive correlations of plasma total homocysteine with DNA methylation were observed at 580 sites (43.3%), and negative correlations were observed at 758 sites (56.7%). Positive correlations were found predominantly at CpG sites in the CGIs. The percentage of the CpG sites with positive correlations, which were located in the CGIs, CGI shores and CGI shelves, were 71.7%, 50.1% and 23.7%, respectively ().
Figure 4. Percentage of CpG sites with positive correlations, located in CGIs, CGI shores and CGI shelves. (A) Of the 212 CpG sites located in the CGIs, 152 (71.7%) showed positive correlations between plasma total homocysteine and DNA methylation. (B) Of the 377 CpG sites located in the CGI shores, 189 (50.1%) showed positive correlations between plasma total homocysteine and DNA methylation. (C) Of the 169 CpG sites located in the CGI shelves, 40 (23.7%) showed positive correlations between plasma total homocysteine and DNA methylation.
Discussion
In this study, we demonstrated that patients with SCZ had significantly elevated plasma total homocysteine levels compared with the controls’ levels, and this result is consistent with the results of a previous meta-analysis.Citation3 We also performed a genome-wide DNA methylation profiling of the peripheral leukocytes in the same subjects with SCZ, and examined the relationship between plasma total homocysteine and DNA methylation patterns. We identified plasma total homocysteine-related changes in DNA methylation at numerous CpG sites. To our knowledge, this is the first study to examine the relationship between plasma total homocysteine and genome-wide DNA methylation in SCZ.
The present study demonstrated that significant correlations between plasma total homocysteine and DNA methylation were observed at CpG sites not only in the promoter regions but also in the gene bodies, and 3′-UTRs. Thus, plasma total homocysteine might affect DNA methylation across whole gene regions. Furthermore, plasma total homocysteine was significantly correlated with DNA methylation at CpG sites not only in the CGIs but also in CGI shores and CGI shelves. Consistent with a previous genome-wide DNA methylation study using cord blood-plasma total homocysteine,Citation32 both positive and negative correlations between plasma total homocysteine and DNA methylation were observed in this study. Notably, the proportions of the CpG sites with positive correlations differed among these three categories (CGI: 71.7%; CGI shore: 50.1%; and CGI shelf: 23.7%). These results suggest that plasma total homocysteine might influence DNA methylation depending on CpG densities.
To date, only one study has examined an association between homocysteine and DNA methylation in patients with SCZ: Bromberg and colleagues measured plasma total homocysteine and global blood DNA methylation in patients with SCZ by using a modification of the radiolabeled [3H]dCTP-extension assay, and they failed to find a significant association.Citation10 This result suggests that DNA methylation must be analyzed at a gene-specific level in studies of SCZ. When we focused on specific genes that demonstrated significant correlations in our study, several genes of these genes, such as SLC18A2, GNAL, KCNH2 and NTNG2, have been implicated in SCZ. SLC18A2 encodes the vesicular transporter type2 (VMAT2), which transports monoamines into the synaptic vesicles.Citation37 Genetic variations of this gene have been associated with SCZ and cognitive functioning in patients with psychotic disorder.Citation34,Citation36,Citation38 GNAL encodes guanine nucleotide-binding protein G subunit α, and altered expression of this gene in the brain is associated with functional changes of dopamine D1 receptor.Citation33 This gene is located in the region of chromosome 18p11.2, and this region has been implicated in susceptibility to bipolar disorder and SCZ.Citation33,Citation35,Citation39 KCNH2 is a member of a family that provides instructions for making potassium channels and that modulates neuronal firing. Altered KCNH2 expression in the hippocampus in SCZ, and a genetic association of this gene with SCZ and SCZ-related neuropsychological deficits in healthy subjects have been reported.Citation40,Citation41 The NTNG2 gene plays a role in synaptic formation and maintenance.Citation42,Citation43 Altered the NTNG2 gene expression in postmortem brains in SCZ and the genetic associations of this gene with SCZ have been reported.Citation44
There are several limitations to the present study. First, the sample size was not large and the risk of potential false-positive results due to multiple testing must be considered. Replication studies with larger samples are necessary. Second, the number of CpG sites that have been analyzed was limited, although the 450K microarray is one of the most powerful tools currently available for assessing DNA methylation changes. Third, the subjects analyzed were chronic patients with SCZ who were receiving treatment with various antipsychotic medications. Antipsychotic drugs are known to influence DNA methylation status.Citation19,Citation20,Citation45,Citation46 Fourth, some genetic variants, clinical symptoms and other components of the methionine cycle, such as S-adenosyl-methionine, folic acid and vitamin B, might be involved in variations of DNA methylation and plasma total homocysteine.Citation21,Citation47-Citation52 Finally, hyperhomocysteinemia has been identified as an independent risk factor for several neurological disorders, such as depression and dementia, in addition to SCZ,Citation3,Citation53-Citation55 so further disease-specific DNA methylation analysis will be necessary.
In summary, significant correlations between plasma total homocysteine and DNA methylation were identified at numerous CpG sites in patients with SCZ, and these results suggest that homocysteine might play a role in the pathogenesis of SCZ via a molecular mechanism involving alterations to DNA methylation.
Materials and Methods
Subjects
Forty-two male patients with SCZ (mean age: 51.8 ± 6.7 y) were recruited from Tokushima and Kochi University Hospitals in Japan. The diagnosis of SCZ was made according to DSM-IV criteria by at least two expert psychiatrists on the basis of extensive clinical interviews and a review of medical records. None of the patients had any psychiatric comorbidity or cardiovascular diseases. All patients were treated with various antipsychotic drugs. The mean chlorpromazine equivalent dose was 829.2 ± 498.2 mg/day. Forty-two male control subjects, well matched for age (mean age: 51.9 ± 5.5 y), were selected from volunteers who were recruited from hospital staff, students and company employees documented to be free from psychiatric problems, past histories of mental illness and medications, including vitamin supplements. All subjects who participated in this study were of unrelated Japanese origin. All subjects signed written informed consent approved by the institutional ethics committees of the University of Tokushima Graduate School and Kochi Medical School.
Plasma total homocysteine analysis
Plasma total homocysteine levels were measured by high performance liquid chromatography. Homocysteine was labeled with 4-fluoro-7-sulfamoylbenzofurazan and detected by a fluorescent detector according to the method of previous studies.Citation10
DNA methylation methods
Genomic DNA was extracted from peripheral blood using the phenol-chloroform method. Bisulfite conversion of 500 ng of genomic DNA was performed with the EZ DNA methylation kit (Zymo Research). DNA methylation level was assessed with Infinium® HumanMethylation450 BeadChips (Illumina Inc.) according to the manufacturer’s instructions. The technical schemes, accuracy and high reproducibility of this array have been described in previous papers.Citation56-Citation58 Quantitative measurements of DNA methylation were determined for 485,764 CpG dinucleotides that covered 99% of the RefSeq genes and were distributed across whole gene regions, including promoters, gene bodies and 3′-UTRs. The arrays also covered 96% of the CGIs from the UCSC database with additional coverage in CGI shores (0–2 kb from CGI) and CGI shelves (2–4 kb from CGI). Detailed information on the contents of the array is available in the Infinium HumanMethylation450 User Guide, HumanMethylation450 manifest (www.illumina.com) and recent papers.Citation56,Citation58 DNA methylation data was analyzed using the methylation analysis module within the BeadStudio software (Illumina Inc.). DNA methylation status of the CpG sites was calculated as the ratio of the signal from a methylated probe relative to the sum of both methylated and unmethylated probes. This value, known as β, ranges from 0 (completely unmethylated) to 1 (fully methylated). For intra-chip normalization of the probe intensities, colored balance and background corrections in every set of 12 samples from the same chip were performed using internal control probes. X chromosome CpG sites in the CGIs in the AR gene as well as the internal control probes were checked to validate the DNA methylation measurements, as in a previous study.Citation59 Of the 485,764 CpG sites, the loci that have β-values of <0.1 or >0.9 were eliminated, as in previous studies.Citation32,Citation60 The loci that are potentially confoundable with single nucleotide polymorphisms with a minor allele frequency of >0.1 in the HapMap-JPT population were also removed because DNA methylation is associated with genotypic variants.Citation61 The final data set includes 164,657 CpG sites.
Statistical methods
Differences in plasma total homocysteine levels between the two groups were examined using a Mann–Whitney U test. The influences of plasma total homocysteine on DNA methylation was examined with a multiple linear regression analysis adjusted for age and chlorpromazine equivalent dose as potential confounders, after standardizing DNA methylation β and plasma total homocysteine values with Z-scores across the samples.
| Abbreviations: | ||
| SCZ | = | Schizophrenia |
| CGI | = | CpG island |
| UTR | = | Untranslated region |
| VMAT2 | = | vesicular transporter type2 |
Additional material
Download Zip (858.6 KB)Acknowledgments
The authors would like to thank all the volunteers, who understood our study purpose and participated in this study, and the physicians, who helped us to collect clinical data and blood samples at the mental hospitals. The authors would also like to thank Mrs. Akemi Okada and Mrs. Kumiko Kikuchi for their technical assistance. The authors also thank Dr. Jörg Tost for his valuable comments and suggestions on SNP-associated probes in the Illumina HumanMethylation450 platform. This work was supported in part by a Grant-in-Aid for Scientific Research from the Japanese Ministry of Education, Culture, Sports, Science and Technology (24791216), SENSHIN Medical Research Foundation and the Research Group For Schizophrenia.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were dislosed.
Supplemental Material
Supplementary materials may be found here:
http://www.landesbioscience.com/journals/epigenetics/article/24621
Notes
† These authors contributed equally to this work.
References
- Saha S, Chant D, Welham J, McGrath J. A systematic review of the prevalence of schizophrenia. PLoS Med 2005; 2:e141; http://dx.doi.org/10.1371/journal.pmed.0020141; PMID: 15916472
- Brown AS, Bottiglieri T, Schaefer CA, Quesenberry CP Jr., Liu L, Bresnahan M, et al. Elevated prenatal homocysteine levels as a risk factor for schizophrenia. Arch Gen Psychiatry 2007; 64:31 - 9; http://dx.doi.org/10.1001/archpsyc.64.1.31; PMID: 17199052
- Muntjewerff JW, Kahn RS, Blom HJ, den Heijer M. Homocysteine, methylenetetrahydrofolate reductase and risk of schizophrenia: a meta-analysis. Mol Psychiatry 2006; 11:143 - 9; http://dx.doi.org/10.1038/sj.mp.4001746; PMID: 16172608
- Dietrich-Muszalska A, Malinowska J, Olas B, Głowacki R, Bald E, Wachowicz B, et al. The oxidative stress may be induced by the elevated homocysteine in schizophrenic patients. Neurochem Res 2012; 37:1057 - 62; http://dx.doi.org/10.1007/s11064-012-0707-3; PMID: 22270909
- Liu CC, Ho WY, Leu KL, Tsai HM, Yang TH. Effects of S-adenosylhomocysteine and homocysteine on DNA damage and cell cytotoxicity in murine hepatic and microglia cell lines. J Biochem Mol Toxicol 2009; 23:349 - 56; http://dx.doi.org/10.1002/jbt.20298; PMID: 19827130
- Kruman II, Culmsee C, Chan SL, Kruman Y, Guo Z, Penix L, et al. Homocysteine elicits a DNA damage response in neurons that promotes apoptosis and hypersensitivity to excitotoxicity. J Neurosci 2000; 20:6920 - 6; PMID: 10995836
- Tyagi N, Moshal KS, Ovechkin AV, Rodriguez W, Steed M, Henderson B, et al. Mitochondrial mechanism of oxidative stress and systemic hypertension in hyperhomocysteinemia. J Cell Biochem 2005; 96:665 - 71; http://dx.doi.org/10.1002/jcb.20578; PMID: 16149054
- Abdolmaleky HM, Cheng KH, Faraone SV, Wilcox M, Glatt SJ, Gao F, et al. Hypomethylation of MB-COMT promoter is a major risk factor for schizophrenia and bipolar disorder. Hum Mol Genet 2006; 15:3132 - 45; http://dx.doi.org/10.1093/hmg/ddl253; PMID: 16984965
- Abdolmaleky HM, Yaqubi S, Papageorgis P, Lambert AW, Ozturk S, Sivaraman V, et al. Epigenetic dysregulation of HTR2A in the brain of patients with schizophrenia and bipolar disorder. Schizophr Res 2011; 129:183 - 90; http://dx.doi.org/10.1016/j.schres.2011.04.007; PMID: 21550210
- Bromberg A, Levine J, Nemetz B, Belmaker RH, Agam G. No association between global leukocyte DNA methylation and homocysteine levels in schizophrenia patients. Schizophr Res 2008; 101:50 - 7; http://dx.doi.org/10.1016/j.schres.2008.01.009; PMID: 18276118
- Carrard A, Salzmann A, Malafosse A, Karege F. Increased DNA methylation status of the serotonin receptor 5HTR1A gene promoter in schizophrenia and bipolar disorder. J Affect Disord 2011; 132:450 - 3; http://dx.doi.org/10.1016/j.jad.2011.03.018; PMID: 21453976
- Chen Y, Zhang J, Zhang L, Shen Y, Xu Q. Effects of MAOA promoter methylation on susceptibility to paranoid schizophrenia. Hum Genet 2012; 131:1081 - 7; http://dx.doi.org/10.1007/s00439-011-1131-5; PMID: 22198720
- Dempster EL, Mill J, Craig IW, Collier DA. The quantification of COMT mRNA in post mortem cerebellum tissue: diagnosis, genotype, methylation and expression. BMC Med Genet 2006; 7:10; http://dx.doi.org/10.1186/1471-2350-7-10; PMID: 16483362
- Dempster EL, Pidsley R, Schalkwyk LC, Owens S, Georgiades A, Kane F, et al. Disease-associated epigenetic changes in monozygotic twins discordant for schizophrenia and bipolar disorder. Hum Mol Genet 2011; 20:4786 - 96; http://dx.doi.org/10.1093/hmg/ddr416; PMID: 21908516
- Ghadirivasfi M, Nohesara S, Ahmadkhaniha HR, Eskandari MR, Mostafavi S, Thiagalingam S, et al. Hypomethylation of the serotonin receptor type-2A Gene (HTR2A) at T102C polymorphic site in DNA derived from the saliva of patients with schizophrenia and bipolar disorder. Am J Med Genet B Neuropsychiatr Genet 2011; 156B:536 - 45; http://dx.doi.org/10.1002/ajmg.b.31192; PMID: 21598376
- Grayson DR, Jia X, Chen Y, Sharma RP, Mitchell CP, Guidotti A, et al. Reelin promoter hypermethylation in schizophrenia. Proc Natl Acad Sci U S A 2005; 102:9341 - 6; http://dx.doi.org/10.1073/pnas.0503736102; PMID: 15961543
- Iwamoto K, Bundo M, Yamada K, Takao H, Iwayama-Shigeno Y, Yoshikawa T, et al. DNA methylation status of SOX10 correlates with its downregulation and oligodendrocyte dysfunction in schizophrenia. J Neurosci 2005; 25:5376 - 81; http://dx.doi.org/10.1523/JNEUROSCI.0766-05.2005; PMID: 15930386
- Kinoshita M, Numata S, Tajima A, Shimodera S, Ono S, Imamura A, et al. DNA methylation signatures of peripheral leukocytes in schizophrenia. Neuromolecular Med 2013; 15:95 - 101; http://dx.doi.org/10.1007/s12017-012-8198-6; PMID: 22961555
- Melas PA, Rogdaki M, Ösby U, Schalling M, Lavebratt C, Ekström TJ. Epigenetic aberrations in leukocytes of patients with schizophrenia: association of global DNA methylation with antipsychotic drug treatment and disease onset. FASEB J 2012; 26:2712 - 8; http://dx.doi.org/10.1096/fj.11-202069; PMID: 22426120
- Mill J, Tang T, Kaminsky Z, Khare T, Yazdanpanah S, Bouchard L, et al. Epigenomic profiling reveals DNA-methylation changes associated with major psychosis. Am J Hum Genet 2008; 82:696 - 711; http://dx.doi.org/10.1016/j.ajhg.2008.01.008; PMID: 18319075
- Nishioka M, Bundo M, Koike S, Takizawa R, Kakiuchi C, Araki T, et al. Comprehensive DNA methylation analysis of peripheral blood cells derived from patients with first-episode schizophrenia. J Hum Genet 2013; 58:91 - 7; http://dx.doi.org/10.1038/jhg.2012.140; PMID: 23235336
- Nohesara S, Ghadirivasfi M, Mostafavi S, Eskandari MR, Ahmadkhaniha H, Thiagalingam S, et al. DNA hypomethylation of MB-COMT promoter in the DNA derived from saliva in schizophrenia and bipolar disorder. J Psychiatr Res 2011; 45:1432 - 8; http://dx.doi.org/10.1016/j.jpsychires.2011.06.013; PMID: 21820670
- Petronis A, Gottesman II, Kan P, Kennedy JL, Basile VS, Paterson AD, et al. Monozygotic twins exhibit numerous epigenetic differences: clues to twin discordance?. Schizophr Bull 2003; 29:169 - 78; http://dx.doi.org/10.1093/oxfordjournals.schbul.a006988; PMID: 12908672
- Pun FW, Zhao C, Lo WS, Ng SK, Tsang SY, Nimgaonkar V, et al. Imprinting in the schizophrenia candidate gene GABRB2 encoding GABA(A) receptor β(2) subunit. Mol Psychiatry 2011; 16:557 - 68; http://dx.doi.org/10.1038/mp.2010.47; PMID: 20404824
- Shimabukuro M, Sasaki T, Imamura A, Tsujita T, Fuke C, Umekage T, et al. Global hypomethylation of peripheral leukocyte DNA in male patients with schizophrenia: a potential link between epigenetics and schizophrenia. J Psychiatr Res 2007; 41:1042 - 6; http://dx.doi.org/10.1016/j.jpsychires.2006.08.006; PMID: 17049557
- Tochigi M, Iwamoto K, Bundo M, Komori A, Sasaki T, Kato N, et al. Methylation status of the reelin promoter region in the brain of schizophrenic patients. Biol Psychiatry 2008; 63:530 - 3; http://dx.doi.org/10.1016/j.biopsych.2007.07.003; PMID: 17870056
- Tolosa A, Sanjuán J, Dagnall AM, Moltó MD, Herrero N, de Frutos R. FOXP2 gene and language impairment in schizophrenia: association and epigenetic studies. BMC Med Genet 2010; 11:114; http://dx.doi.org/10.1186/1471-2350-11-114; PMID: 20649982
- Bönsch D, Lenz B, Reulbach U, Kornhuber J, Bleich S. Homocysteine associated genomic DNA hypermethylation in patients with chronic alcoholism. J Neural Transm 2004; 111:1611 - 6; http://dx.doi.org/10.1007/s00702-004-0232-x; PMID: 15565495
- Huang YS, Zhi YF, Wang SR. Hypermethylation of estrogen receptor-α gene in atheromatosis patients and its correlation with homocysteine. Pathophysiology 2009; 16:259 - 65; http://dx.doi.org/10.1016/j.pathophys.2009.02.010; PMID: 19285843
- Ingrosso D, Cimmino A, Perna AF, Masella L, De Santo NG, De Bonis ML, et al. Folate treatment and unbalanced methylation and changes of allelic expression induced by hyperhomocysteinaemia in patients with uraemia. Lancet 2003; 361:1693 - 9; http://dx.doi.org/10.1016/S0140-6736(03)13372-7; PMID: 12767735
- Lv H, Ma X, Che T, Chen Y. Methylation of the promoter A of estrogen receptor alpha gene in hBMSC and osteoblasts and its correlation with homocysteine. Mol Cell Biochem 2011; 355:35 - 45; http://dx.doi.org/10.1007/s11010-011-0836-z; PMID: 21523370
- Fryer AA, Emes RD, Ismail KM, Haworth KE, Mein C, Carroll WD, et al. Quantitative, high-resolution epigenetic profiling of CpG loci identifies associations with cord blood plasma homocysteine and birth weight in humans. Epigenetics 2011; 6:86 - 94; http://dx.doi.org/10.4161/epi.6.1.13392; PMID: 20864804
- Corradi JP, Ravyn V, Robbins AK, Hagan KW, Peters MF, Bostwick R, et al. Alternative transcripts and evidence of imprinting of GNAL on 18p11.2. Mol Psychiatry 2005; 10:1017 - 25; http://dx.doi.org/10.1038/sj.mp.4001713; PMID: 16044173
- Gutiérrez B, Rosa A, Papiol S, Arrufat FJ, Catalán R, Salgado P, et al. Identification of two risk haplotypes for schizophrenia and bipolar disorder in the synaptic vesicle monoamine transporter gene (SVMT). Am J Med Genet B Neuropsychiatr Genet 2007; 144B:502 - 7; http://dx.doi.org/10.1002/ajmg.b.30499; PMID: 17427184
- Schwab SG, Hallmayer J, Lerer B, Albus M, Borrmann M, Hönig S, et al. Support for a chromosome 18p locus conferring susceptibility to functional psychoses in families with schizophrenia, by association and linkage analysis. Am J Hum Genet 1998; 63:1139 - 52; http://dx.doi.org/10.1086/302046; PMID: 9758604
- Talkowski ME, Kirov G, Bamne M, Georgieva L, Torres G, Mansour H, et al. A network of dopaminergic gene variations implicated as risk factors for schizophrenia. Hum Mol Genet 2008; 17:747 - 58; http://dx.doi.org/10.1093/hmg/ddm347; PMID: 18045777
- Taylor SF, Koeppe RA, Tandon R, Zubieta JK, Frey KA. In vivo measurement of the vesicular monoamine transporter in schizophrenia. Neuropsychopharmacology 2000; 23:667 - 75; http://dx.doi.org/10.1016/S0893-133X(00)00165-2; PMID: 11063922
- Simons CJ, van Winkel R, GROUP. Intermediate phenotype analysis of patients, unaffected siblings, and healthy controls identifies VMAT2 as a candidate gene for psychotic disorder and neurocognition. Schizophr Bull 2012; http://dx.doi.org/10.1093/schbul/sbs067; PMID: 22532702
- Stine OC, Xu J, Koskela R, McMahon FJ, Gschwend M, Friddle C, et al. Evidence for linkage of bipolar disorder to chromosome 18 with a parent-of-origin effect. Am J Hum Genet 1995; 57:1384 - 94; PMID: 8533768
- Hashimoto R, Ohi K, Yasuda Y, Fukumoto M, Yamamori H, Kamino K, et al. The KCNH2 gene is associated with neurocognition and the risk of schizophrenia. World J Biol Psychiatry 2013; 14:114 - 20; http://dx.doi.org/10.3109/15622975.2011.604350; PMID: 21936766
- Huffaker SJ, Chen J, Nicodemus KK, Sambataro F, Yang F, Mattay V, et al. A primate-specific, brain isoform of KCNH2 affects cortical physiology, cognition, neuronal repolarization and risk of schizophrenia. Nat Med 2009; 15:509 - 18; http://dx.doi.org/10.1038/nm.1962; PMID: 19412172
- Eastwood SL, Harrison PJ. Decreased mRNA expression of netrin-G1 and netrin-G2 in the temporal lobe in schizophrenia and bipolar disorder. Neuropsychopharmacology 2008; 33:933 - 45; http://dx.doi.org/10.1038/sj.npp.1301457; PMID: 17507910
- Eastwood SL, Harrison PJ. Markers of glutamate synaptic transmission and plasticity are increased in the anterior cingulate cortex in bipolar disorder. Biol Psychiatry 2010; 67:1010 - 6; http://dx.doi.org/10.1016/j.biopsych.2009.12.004; PMID: 20079890
- Aoki-Suzuki M, Yamada K, Meerabux J, Iwayama-Shigeno Y, Ohba H, Iwamoto K, et al. A family-based association study and gene expression analyses of netrin-G1 and -G2 genes in schizophrenia. Biol Psychiatry 2005; 57:382 - 93; http://dx.doi.org/10.1016/j.biopsych.2004.11.022; PMID: 15705354
- Shimabukuro M, Jinno Y, Fuke C, Okazaki Y. Haloperidol treatment induces tissue- and sex-specific changes in DNA methylation: a control study using rats. Behav Brain Funct 2006; 2:37; http://dx.doi.org/10.1186/1744-9081-2-37; PMID: 17132176
- Tremolizzo L, Doueiri MS, Dong E, Grayson DR, Davis J, Pinna G, et al. Valproate corrects the schizophrenia-like epigenetic behavioral modifications induced by methionine in mice. Biol Psychiatry 2005; 57:500 - 9; http://dx.doi.org/10.1016/j.biopsych.2004.11.046; PMID: 15737665
- Krebs MO, Bellon A, Mainguy G, Jay TM, Frieling H. One-carbon metabolism and schizophrenia: current challenges and future directions. Trends Mol Med 2009; 15:562 - 70; http://dx.doi.org/10.1016/j.molmed.2009.10.001; PMID: 19896901
- Ulrey CL, Liu L, Andrews LG, Tollefsbol TO. The impact of metabolism on DNA methylation. Hum Mol Genet 2005; 14:Spec No 1 R139 - 47; http://dx.doi.org/10.1093/hmg/ddi100; PMID: 15809266
- Bouaziz N, Ayedi I, Sidhom O, Kallel A, Rafrafi R, Jomaa R, et al. Plasma homocysteine in schizophrenia: determinants and clinical correlations in Tunisian patients free from antipsychotics. Psychiatry Res 2010; 179:24 - 9; http://dx.doi.org/10.1016/j.psychres.2010.04.008; PMID: 20471108
- Hazra A, Kraft P, Lazarus R, Chen C, Chanock SJ, Jacques P, et al. Genome-wide significant predictors of metabolites in the one-carbon metabolism pathway. Hum Mol Genet 2009; 18:4677 - 87; http://dx.doi.org/10.1093/hmg/ddp428; PMID: 19744961
- Paré G, Chasman DI, Parker AN, Zee RR, Mälarstig A, Seedorf U, et al. Novel associations of CPS1, MUT, NOX4, and DPEP1 with plasma homocysteine in a healthy population: a genome-wide evaluation of 13 974 participants in the Women’s Genome Health Study. Circ Cardiovasc Genet 2009; 2:142 - 50; http://dx.doi.org/10.1161/CIRCGENETICS.108.829804; PMID: 20031578
- Tanaka T, Scheet P, Giusti B, Bandinelli S, Piras MG, Usala G, et al. Genome-wide association study of vitamin B6, vitamin B12, folate, and homocysteine blood concentrations. Am J Hum Genet 2009; 84:477 - 82; http://dx.doi.org/10.1016/j.ajhg.2009.02.011; PMID: 19303062
- Almeida OP, McCaul K, Hankey GJ, Norman P, Jamrozik K, Flicker L. Homocysteine and depression in later life. Arch Gen Psychiatry 2008; 65:1286 - 94; http://dx.doi.org/10.1001/archpsyc.65.11.1286; PMID: 18981340
- Bjelland I, Tell GS, Vollset SE, Refsum H, Ueland PM. Folate, vitamin B12, homocysteine, and the MTHFR 677C->T polymorphism in anxiety and depression: the Hordaland Homocysteine Study. Arch Gen Psychiatry 2003; 60:618 - 26; http://dx.doi.org/10.1001/archpsyc.60.6.618; PMID: 12796225
- Wald DS, Kasturiratne A, Simmonds M. Serum homocysteine and dementia: meta-analysis of eight cohort studies including 8669 participants. Alzheimers Dement 2011; 7:412 - 7; http://dx.doi.org/10.1016/j.jalz.2010.08.234; PMID: 21784352
- Bibikova M, Barnes B, Tsan C, Ho V, Klotzle B, Le JM, et al. High density DNA methylation array with single CpG site resolution. Genomics 2011; 98:288 - 95; http://dx.doi.org/10.1016/j.ygeno.2011.07.007; PMID: 21839163
- Dedeurwaerder S, Defrance M, Calonne E, Denis H, Sotiriou C, Fuks F. Evaluation of the Infinium Methylation 450K technology. Epigenomics 2011; 3:771 - 84; http://dx.doi.org/10.2217/epi.11.105; PMID: 22126295
- Sandoval J, Heyn H, Moran S, Serra-Musach J, Pujana MA, Bibikova M, et al. Validation of a DNA methylation microarray for 450,000 CpG sites in the human genome. Epigenetics 2011; 6:692 - 702; http://dx.doi.org/10.4161/epi.6.6.16196; PMID: 21593595
- Siegmund KD, Connor CM, Campan M, Long TI, Weisenberger DJ, Biniszkiewicz D, et al. DNA methylation in the human cerebral cortex is dynamically regulated throughout the life span and involves differentiated neurons. PLoS One 2007; 2:e895; http://dx.doi.org/10.1371/journal.pone.0000895; PMID: 17878930
- Byun HM, Siegmund KD, Pan F, Weisenberger DJ, Kanel G, Laird PW, et al. Epigenetic profiling of somatic tissues from human autopsy specimens identifies tissue- and individual-specific DNA methylation patterns. Hum Mol Genet 2009; 18:4808 - 17; http://dx.doi.org/10.1093/hmg/ddp445; PMID: 19776032
- Numata S, Ye T, Hyde TM, Guitart-Navarro X, Tao R, Wininger M, et al. DNA methylation signatures in development and aging of the human prefrontal cortex. Am J Hum Genet 2012; 90:260 - 72; http://dx.doi.org/10.1016/j.ajhg.2011.12.020; PMID: 22305529