Abstract
Recently, it has been suggested that C2ORF40 is a candidate tumor suppressor gene in breast cancer. However, the mechanism for reduced expression of C2ORF40 and its functional role in breast cancers remain unclear. Here we show that C2ORF40 is frequently silenced in human primary breast cancers and cell lines through promoter hypermethylation. C2ORF40 mRNA level is significantly associated with patient disease-free survival and distant cancer metastasis. Overexpression of C2ORF40 inhibits breast cancer cell proliferation, migration and invasion. By contrast, silencing C2ORF40 expression promotes these biological phenotypes. Bioinformatics and FACS analysis reveal C2ORF40 functions at G2/M phase by downregulation of mitotic genes expression, including UBE2C. Our results suggest that C2ORF40 acts as a tumor suppressor gene in breast cancer pathogenesis and progression and is a candidate prognostic marker for this disease.
Introduction
Breast cancers are the most frequently diagnosed cancer and the second leading cause of cancer-related death among women in both economically developed and developing countries, accounting for 23% of the total cancer cases and 14% of the cancer deaths.Citation1 Despite research endeavors and resources dedicated to elucidating the molecular mechanisms of breast cancers, and numerous genetic variants and genes with irregular expression discovered over the past decades,Citation2 the precise molecular mechanisms of initiation and progression of this heterogeneous cancer still remain largely unclear. This ambiguity hampers the design of efficient and personalized chemotherapy and biotherapy strategies. Thus, finding new breast cancer-related genes and elucidating their function and clinical implication in breast cancers are urgently demanded.
During our efforts to discover new novel targets significantly associated with breast cancer patient prognosis by integrative analysis of existing public data, we found that C2ORF40 (chromosome 2 open reading frame 40, also called esophageal cancer-related gene 4 (ECRG4) or augurin) is one at the top of the list of genes, the transcriptional levels of which are dramatically reduced in breast cancers and are significantly correlated with patient survival.
C2ORF40 was initially identified and cloned from human normal esophageal epithelium by comparing differential gene expression between normal human esophageal epithelia and ESCC from high incidence families in China.Citation3 Although the expression of C2ORF40 was ubiquitously detected in normal tissues,Citation4,Citation5 it is frequently downregulated or absent in esophageal cancer, colorectal carcinomas and glioma, probably due to promoter hypermethylation,Citation6,Citation7 and its expression is thought to be a prognostic factor for ESCC and prostate cancer patients.Citation8,Citation9 Restoration of C2ORF40 expression in cell lines could inhibit esophageal, colorectal and glioma tumor cell growthCitation7,Citation9-Citation11 and glioma cell migration.Citation10 Together with these reports, our preliminary results obtained by analysis of gene expression database suggest that C2ORF40 may also play a tumor suppressor role in breast cancers. Recently, one study suggested that C2ORF40 is a candidate tumor suppressor gene in breast cancer.Citation12 However, the mechanism for reduced expression of C2ORF40 and its functional role in breast cancers have not been reported.
In this study, we first evaluated C2ORF40 expression and its clinical prognostic significance in breast cancer patients. Then, we examined the possibility that C2ORF40 was epigenetically inactivated through promoter hypermethylation in human breast cancer cell lines and primary breast cancer tissues. Further, we investigated the tumor suppressing function of C2ORF40 in terms of cell proliferation, migration and invasion in breast cancer cells. Finally we explored the possible molecular mechanisms through which C2ORF40 might be involved in suppression of breast cancers. The present work is part of our effort to understand the linkage between gene expression profiles and breast cancer outcomes in order to find new diagnostic and prognostic biomarkers and new molecular therapy targets.
Results
Loss of C2ORF40 expression associates with poor prognosis in breast cancer patients
We first analyzed C2ORF40 expression in primary breast cancer and normal breast tissues using all gene expression profile data sets that can be found in the Gene Expression Omnibus (GEO) database (Table S1). Of 100 breast cancer samples and 164 normal breast tissues in Data set 1 to 3, C2ORF40 transcript level in tumors is dramatically and consistently reduced compared with normal breast tissues ().
Figure 1. Reduced C2ORF40 mRNA expression was a significant prognostic factor for disease-free and metastasis-free survival. C2ORF40 expression assessed by Affymetrix microarray in human breast cancers and normal tissues in data set 1 to 3 (Table S1) was shown in panel (A) to (C). The significant association between C2ORF40 mRNA level and disease-free survival was analyzed in four independent cohorts of breast cancer patients (Data set 4 to 7 in Table S1) (D) to (G). The patients from each cohort were divided into groups with high (top one-third), moderate (middle one-third) and low (bottom one-third) level of C2ORF40 expression. Panel (D) to (G) show the Kaplan-Meier survival curves for disease-free survival in the four data sets respectively. Panels (H) and (I) show the Kaplan-Meier survival curves for metastasis-free survival in Data set 4 (NKI) and Data set 7 (GSE6532) respectively. C2ORF40 mRNA is measured as log2 (probe intensities) as in the microarray. The P-values shown were obtained from Mann-Whitney U (A and B), Kruskal-Wallis (C) or long-rank tests (D to I).
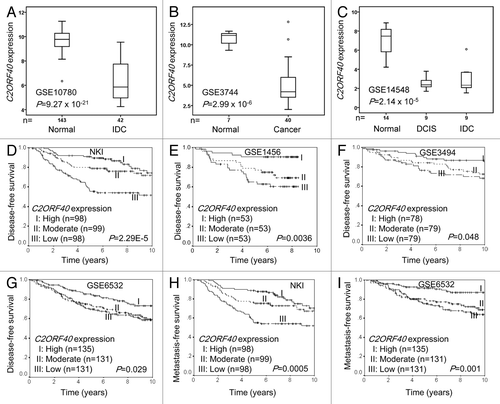
To evaluate the clinical implication of reduction of C2ORF40 expression in breast cancers, we assessed the association of C2ORF40 mRNA levels with breast cancer patient survival using all GEO microarray data sets of primary breast cancer patients with available clinical information (Table S1). When C2ORF40 expression levels were divided into three groups (low = bottom third, moderate = middle third and high = top third), we found, in all of 4 data sets (Data set 4 to 7 in Table S1), patients with lower C2ORF40 mRNA levels had significantly shorter disease-free survival (). In addition, analyses of two independent cohorts of breast cancer patients with available distant metastasis information showed that patients with lower C2ORF40 mRNA levels had more chances of distant metastasis in comparison to patients with higher level of C2ORF40 expression (). All these results clearly indicate that C2ORF40 is a potential tumor suppressor gene and its mRNA level is a potential prognostic biomarker for breast cancers.
Reduction of C2ORF40 expression in breast cancers is due to promoter hypermethylation
To elucidate possible mechanisms mediating the loss of C2ORF40 expression in breast cancers, we first examined genomic aberrations in C2ORF40 gene locus by analyzing the previously reported comparative genomic hybridization (CGH) data of 130 primary breast cancersCitation13 and 54 human breast cancer cell lines,Citation14 and observed no frequent genetic alterations in C2ORF40 locus (Fig. S1), such as deletion or gain determined as described by ChinCitation13 and Neve,Citation14 which led us to hypothesize that the alteration of C2ORF40 expression might be due to epigenetic modification, since hypermethylation of CpG islands of gene promoter often causes transcriptional silencing of tumor suppressor genes. To clarify whether loss of C2ORF40 expression was due to hypermethylation of its promoter, we analyzed the sequence 0.42 kb upstream and 0.22 kb downstream of the transcription initiation site of C2ORF40 gene. Sixty-one CpG sites were identified throughout this region () and the methylation status of each of these CpGs was examined in 22 cell lines by bisulfite sequence analysis and methylation specific PCR (). C2ORF40 promoter was hypermethylated in 45% (10 out of 22) of these breast cancer cell lines (). Moreover, promoter hypermethylation status in these cell lines was correlated with its expression (). Selected cell lines were then treated with demethylation drug 5-aza-dC. 5-aza-dC treatment significantly increased C2ORF40 mRNA levels in hypermethylated BT549 and MDAMB231 and partially hypermethylated AU565 cell lines (). In contrast, C2ORF40 expression in cell lines such as HCC70 and SKBR3 without C2ROF40 promoter hypermethylation did not change after 5-aza-dC treatment (). Next, we examined whether C2ORF40 is methylated in human primary breast cancers by methylation specific PCR. Indeed, C2ORF40 promoter was hypermethylated in 38.5% (37 out of 96) of primary breast cancer tissues we collected. Furthermore, this hypermethylation status was tightly correlated with C2ORF40 expression (). These results clearly demonstrated that promoter hypermethylation could be the main mechanism leading to silencing of C2ORF40 gene in breast cancers.
Figure 2. Loss of C2ORF40 expression due to promoter hypermethylation. (A) Schematic map of the 5′-CpG islands located in the C2ORF40 promoter region around the transcription start site (ATG). CpG dinucleotides are depicted. PCR primers used for sodium bisulfite sequencing (SF1, SF2, SR1 and SR2) and for detecting methylation (MF and MR) and non-methylation (UF and UR) are indicated as arrows. The sequences of these primers are listed in Materials and Methods. (B) The representative sequence traces show the methylation of C2ORF40 promoter. (C) Analysis of C2ORF40 promoter methylation using MSPCR. U indicates unmethylated, M indicates methylated. (D) The relationship between C2ORF40 expression and its promoter methylation in a panel of human breast cancer cell lines. (E) qRT-PCR shows C2ORF40 expression is lower in promoter hypermethylated MDAMB231and BT549 cells than in unmethylated SKBR3, HCC70 cell lines and 5-aza-dC treatment restored C2ORF40 expression in MDAMB231 and BT549 cell lines. (F) The relationship between C2ORF40 expression and its promoter methylation in human primary breast cancers. Each experiment in (B), (C) and (E) was repeated at least three times. Data were presented as boxplot based on C2ORF40 promoter methylation status in (D) and (F) and mean ± SD in (E). The P-values shown in (D) and (F) were obtained from nonparametric test. * p < 0.05 and ** p < 0.01 in (E) obtained from Student’s t-test as compared with control groups.
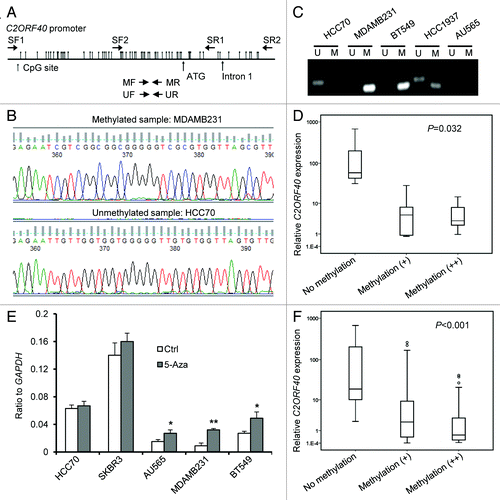
Table 1.C2ORF40 promoter methylation status in human breast cancer cell lines
Restoration of C2ORF40 expression inhibits breast cancer cell growth and invasion
To clarify whether C2ORF40 functions as a tumor suppressor in breast cancer pathogenesis and progression, we examined the effect of C2ORF40 on cell proliferation and migration-invasion by ectopic expression of C2ORF40 in breast cancer cell lines. BT549 and MDAMB231 cell lines were selected based on that C2ORF40 promoter was hypermethylated and its expression was low in these two cell lines ( and ). Stable restoration of C2ORF40 expression was achieved in these two cell lines by retroviral transduction (). As evidenced by MTT assays, proliferation rates of BT549 and MDAMB231 cells with forced expression of C2ORF40 were significantly reduced to 43.7% and 63.7% respectively in comparison to their control cells (). Moreover, BT549 and MDAMB231 cells with overexpression of C2ORF40 formed fewer colonies than their controls ().
Figure 3. Ectopic expression of C2ORF40 inhibits breast cancer cell growth and migration-invasion. (A) C2ORF40 is stably overexpressed in retroviral transduced BT549 and MDAMB231 cells as indicated by RT-PCR (upper panel) and western blotting (lower panel). Overexpression of C2ORF40 inhibits BT549 and MDAMB231 cell proliferation assessed by MTT method (B, C) and colony formation assay (D, E). (B, D) for BT549 and (C, E) for MDAMB231 cells, respectively. Representative photographs show ectopic expression of C2ORF40 inhibits migration of BT549 (F) and MDAMB231 (G) cells in a scratch-wound healing model on cultured cells. Ectopic expression of C2ORF40 in BT549 (H) and MDAMB231 (I) cells decreases cancer cell invasive abilities using Matrigel assay. All experiments were repeated at least three times in two independently retrovirally transduced cell lines. Data were presented as mean ± SD, * p < 0.05 and ** p < 0.01 obtained from Student’s t-test as compared with control groups. Ctrl: empty vector infected control groups.
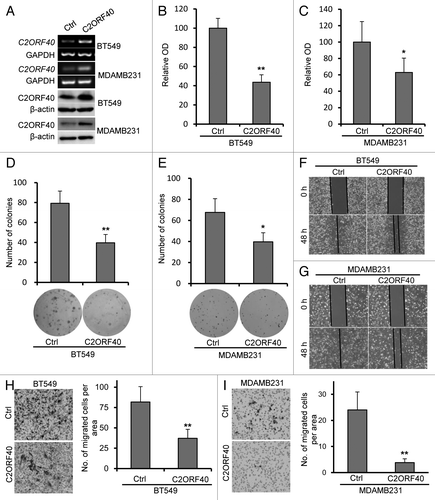
We next examined whether C2ORF40 could modulate the metastatic capacities of breast cancer cells. Confluent BT549 and MDAMB231 cells with or without forced C2ORF40 expression were scratched and cell migration was observed. BT549 and MDAMB231 cells with ectopic expression of C2ORF40 statistically significantly delayed closure of wound area compared with their own control cells (). Their invasive ability was assessed using Matrigel assay. Cells growing in the log phase were collected and cultured in Boyden chamber with Matrigel. After 16 h incubation, BT549 and MDAMB231 cells with C2ORF40 showed a significant decrease in invasiveness, compared with their own controls (). These results clearly demonstrated that restoration of C2ORF40 expression inhibits the migration and invasion of breast cancer cells.
Knockdown of C2ORF40 expression promotes breast cancer cell growth and invasion
The findings described above implicate C2ORF40 as breast cancer suppressor gene. We next further investigated the biological consequences of silencing of C2ORF40 in breast cancer cells. We designed two C2ORF40 specific short hairpin RNA (designated as shA and shB respectively) and generated stable transfectants in SKBR3 and MDAMB468 cells. As shown in , both shA and shB significantly reduced expression of C2ORF40, but shA knocked down C2ORF40 more efficiently than shB. A scrambled shRNA was served as a control (shCtrl). SKBR3 and MDAMB468 cells with shA exhibited a consistent and significant increase in cell proliferation, whereas SKBR3 and MDAMB468 cells with shB had less degree (), indicating dose-dependent effect of C2ORF40 on cell proliferation.
Figure 4. Knockdown of C2ORF40 promotes breast cancer cell growth and migration-invasion. (A) Knockdown of C2ORF40 in SKBR3 and MDAMB468 cells as indicated by RT-PCR (upper panel) and western blotting (lower panel). Knockdown of C2ORF40 promotes SKBR3 and MDAMB468 cell proliferation assessed by MTT method (B, C). Representative photographs show knockdown of C2ORF40 promotes migration of SKBR3 (D) and MDAMB468 (E) cells in a scratch-wound healing model on cultured cells. Knockdown of C2ORF40 promotes the invasive ability of SKBR3 (F) and MDAMB468 (G) cells using Matrigel assay. Images in (D) to (G) show representative field of views. All experiments were repeated at least three times. Data were presented as mean ± SD, * p < 0.05 and ** p < 0.01 obtained from Student’s t-test as compared with control groups. shCtrl: scrambled vector infected control groups.
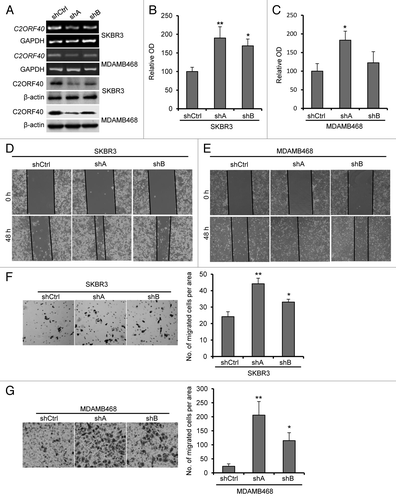
Next, we investigated the effect of C2ORF40 knockdown on cell migration using a wound-healing assay. As shown in , SKBR3 and MDAMB468 cells with C2ORF40 knockdown had faster closure of the wound area compared with their control. The inhibitory effect of C2ORF40 on cell invasion was confirmed using Matrigel assay, in which the SKBR3 and MDAMB468 cells with C2ORF40 knockdown showed a significant increase in cell invasion through Matrigel than their control cells (). Again, we found a dose-dependent effect of C2ORF40 on cell migration and invasion since shA and shB showed some differences (). Taken together, these results clearly indicate that knockdown of C2ORF40 increases the proliferation and the migration-invasion of breast cancer cells.
C2ORF40 suppresses mitosis and mitosis-associated gene expression in breast cancers
In attempts to explore the molecular mechanisms that C2ORF40 regulates breast cancer progression, the genes whose expressions were correlated with the downregulation of C2ORF40 expression were searched in three data sets (GSE1456, GSE3494 and GSE6532, Table S1). Selection of these three data sets was due to all data generated from the same platform of Affymetrix microarray, which allowed us to define the core list of genes among them. This process defined a network comprised of 918 Affymetrix probes representing 693 known genes that reach the criteria (absolute correlation coefficient ≥ 0.3 and FDR < 0.01 in all three data set) (Table S2). IPA analysis of the 693 genes revealed enrichment for genes involved in cell cycle, cellular assembly and organization, DNA replication, recombination and repair () and cellular movement, cancer, cardiovascular system development and function (). These are in agreement with the above biological findings that C2ORF40 inhibits cell proliferation and cell migration-invasion. Gene ontology (GO) analysis revealed that the top-ranked GO terms are involved in mitotic processes (, Table S3).
Figure 5.C2ORF40 regulates mitosis-associated gene expression and M phase progression of cell cycle. IPA networks of genes significantly correlated with C2ORF40 invoked cell cycle, cellular assembly and organization, DNA replication, recombination and repair (A) and cellular movement, cancer, cardiovascular system development and function (B). GO terms associated with lists of genes that are significantly correlated with C2ORF40(C). Cell cycle progression in control vector (left panels) and C2ORF40 (right panels) transfected BT549 (D) and MDAMB231 (E) cells was determined by FACS Caliber cytometry. There is a significant increase in G2/M cell population in the cells with enforced expression of C2ORF40(F). The experiments in (D to F) were repeated at least three times in two independently retrovirally transduced cell lines and data in (F) were presented as mean ± SD P-values shown in (F) were obtained from Student’s t-test. * p < 0.05 and ** < 0.01. Ctrl: empty vector infected control groups.
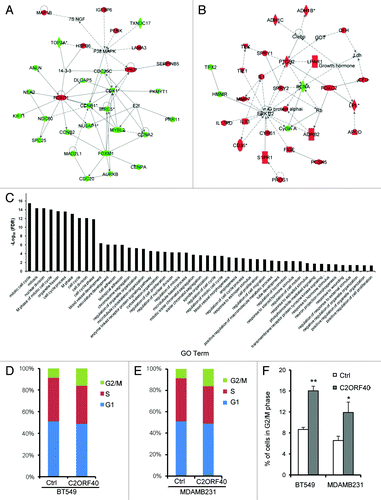
In order to confirm the role of C2ORF40 in mitotic process, cell cycle analysis was performed by flow cytometry. Indeed, forced expression of C2ORF40 arrested breast cancer cells at G2/M phase of cell cycle since the G2/M population in both BT549 and MDAMB231 cells with C2ORF40 was significantly increased in comparison to their respective control cells ().
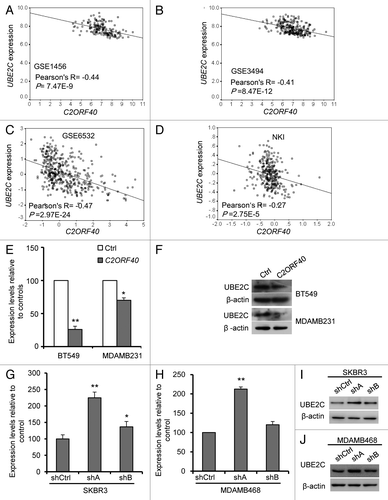
Ubiquitin-conjugating enzyme E2C (UBE2C), an important component of anaphase-promoting complex/cyclosome (APC/c), was significantly and negatively correlated with C2ORF40 expression in the four data sets we analyzed (, Table S2). To further conform that C2ORF40 regulates UBE2C expression, we examined UBE2C expression in the cells with ectopic expression and knockdown of C2ORF40. Restoration of C2ORF40 in BT549 and MDAMB231 cells markedly decreased the expression levels of UBE2C (). In contrast, knockdown of C2ORF40 in SKBR3 and MDAMB468 cells significantly increased the expression of UBE2C (). Taken together, our data clearly suggested that C2ORF40 might block cell cycle progression at M phase through inhibiting mitosis promoting gene expression such as UBE2C.
Figure 6.C2ORF40 suppresses UBE2C expression. There is a highly significant and negative correlation between C2ORF40 and UBE2C in mRNA levels within human breast cancer tissues from GEO database Data set 5 (A), Data set 6 (B), Data set 7 (C), Data set 4 (D). Ectopic expression of C2ORF40 in BT549 and MDAMB231 cells downregulates the mRNA (E) and protein (F) levels of UBE2C. Knockdown of C2ORF40 in SKBR3 (G and I) and MDAMB468 (H and J) cells increases the mRNA (G and H) and protein (I and J) levels of UBE2C. The experiments in (E) to (J) were repeated at least three times and data were presented as mean ± SD. R is Pearson correlation coefficient. P-values shown in (A to D) were obtained from Pearson correlation test and in (E), (G) and (H) were obtained from Student’s t-test. * p < 0.05 and ** < 0.01. Ctrl: empty vector infected control groups, shCtrl: scrambled vector infected control groups.
Discussion
We were prompted to study C2ORF40 gene because it repeatedly appeared on the top of the list of genes whose expression are associated with breast cancer patient survival when we reanalyzed publicly accessible microarray data sets of primary breast cancers previously deposited in GEO database. Our data not only demonstrate that C2ORF40 expression is downregulated in breast cancers and is highly correlated with patient disease-free survival, which is consistent with a recent study showing the clinical implication of C2ORF40 in breast cancers,Citation12 but also show that C2ORF40 expression is highly correlated with breast cancer metastasis, which is further supported by our biological studies. The downregulation of C2ORF40 in breast cancer is mainly due to its promoter hypermethylation. Furthermore, C2ORF40, as a potential tumor suppressor, inhibits cell proliferation and migration-invasion by blocking cell cycle progression at G2/M phase through suppressing mitosis-regulating genes such as UBE2C.
Human breast cancers are heterogeneous in pathologies and molecular profiles. Thus, breast cancer patients have different prognostic outcomes. The current clinical staging systems and molecular classification for breast cancers have limitations in predicting clinical prognosis.Citation15 Finding new prognostic markers has important clinical implications in identifying breast cancer patients at high risk of metastasis and in predicting postsurgical recurrence or survival of breast cancer patients. Consistent with that C2ORF40 has been proposed to be an independent prognostic factor for ESCC patientsCitation8,Citation9; we confirmed that this gene is a potential prognostic marker for breast cancers in this study, which has been suggested by a recent study.Citation12 C2ORF40 mRNA level was analyzed in 3 independent cohorts of microarray data sets including total 164 normal breast tissues and 100 breast tumors and was consistently found lower in tumors than in normal tissues. Furthermore, Kaplan-Meier analysis of more than 1,000 cases of breast cancers in four different cohorts of patients indicated that the disease-free survival and metastasis-free survival of breast cancer patients are associated with C2ORF40 expression levels. These analyses suggest that downregulation of C2ORF40 levels might be an index for breast cancer patient outcomes. Further detailed analyses are required to validate the clinical utility of C2ORF40 as a prognostic biomarker for breast cancers.
The downregulation of tumor suppressor gene expression in cancers could be caused by genetic mutations, loss of heterozygosity (LOH) or epigenetic modification. We analyzed the existing CGH data and found few genomic alterations in C2ORF40 loci in breast cancer, suggesting DNA loss is not main mechanism for downregulation of C2ORF40 expression, which is consistent with the recent report.Citation12 We then analyzed methylation status of total 61 CpG sites around the transcriptional start site. Frequent methylation of these CpG sites was found in primary cancer tissues and commonly used cell lines.Citation16 Furthermore, this hypermethylation strongly correlates with expression of C2ORF40 in breast cancers. C2ORF40 expression could be reactivated by demethylating treatment, which is a known feature of epigenetically silenced tumor suppressor genes.Citation17 In addition, copy number analysis of C2ORF40 gene by qPCR found that only 7 cancers have deletion in our 96 primary breast cancer samples (data not shown). Therefore, we, for the first time, demonstrate that promoter hypermethylation is the main mechanism for loss of C2ORF40 gene expression since few genomic alterations of C2ORF40 gene are found in human breast cancers.
DNA hypermethylation constitutes a major cause of abnormal gene silencing in cancers.Citation18 However, not all aberrant gene silencing plays functional roles in tumor development, some methylation events might be epigenetic passengers.Citation19 Therefore, each candidate tumor suppressor gene identified by methylation and bioinformatics analysis methods has to be carefully evaluated for its functional roles in tumor development and progression. To examine whether C2ORF40 plays a suppressive role in breast cancers, we restored C2ORF40 expression in C2ORF40 promoter hypermethylated breast cancer cells to investigate its biological function. Subsequent MTT and colony formation assays demonstrated that overexpression of C2ORF40 led to significant decrease in cell growth in vitro. Inhibitory effects of C2ORF40 on migration-invasion were also evidenced. Silencing C2ORF40 expression promoted breast cancer cell proliferation, migration and invasion. These are consistent with the findings by Li et al.Citation9,Citation11 that upregulation of C2ORF40 inhibits cell proliferation and invasiveness in ESCC and our bioinformatics analysis of clinical information of breast cancer patients. Our data provided the first biological evidence that C2ORF40 may serve as a tumor suppressor in breast cancers.
Cell cycle analysis further indicated that C2ORF40 might act as a tumor suppressor gene in breast cancers by inhibiting tumor cell growth through inducing cell cycle arrest at G2/M phase, which differs with previous reports in other types of cancers such as ESCC in which C2ORF40 was reported to block G1/S transition.Citation9,Citation20 We do not have any clues for this disparity at this moment; the intrinsic differences in each type of tumors might be a possible explanation. Supporting our cell cycle analysis, more multinucleated cells were observed in C2ORF40-overexpressing cell lines.
C2ORF40 target genes still remain discovery. Correlation of gene expression from more than 1000 primary breast cancer cases indicated that half of the genes whose expressions are negatively correlated with C2ORF40 expression are M-phase promoting factors. This further implied that C2ORF40 might be mainly related with mitosis regulation. Among these M-phase promoting genes, UBE2C is of particular interest as the expression of this gene was found to be critical during spindle assembly, cytokinesis and progression through mitosis,Citation21,Citation22 and loss of UBE2C leads to severe spindle defects and a strong mitotic delay, arresting cell cycle at G2/M phase.Citation22,Citation23 Thus, we speculated C2ORF40 might suppress breast cancer cell growth and block G2/M phase progression by regulating the UBE2C expression. Consistent with this hypothesis, we showed that downregulation of C2ORF40 is negatively correlated UBE2C expression in more than 1,000 primary breast cancer tissues from five independent cohorts of patients and overexpression of C2ORF40 in breast cancer cells markedly suppresses UBE2C expression.
As C2ORF40 gene product was reported as a secretary moleculeCitation5 and can be detected in cell culture medium,Citation7 it is very likely that C2ORF40 exerts its inhibitory function through binding to cell membrane surface molecules to transduce inhibitory signals into adjacent cells.Citation20 Moreover, further processing of this 148-amino acid molecule into smaller peptides is reportedly required for its inhibitory functionCitation24 and smaller processed peptides are found in cell culture medium.Citation25,Citation26 Thus, C2ORF40 is not only a prognostic biomarker and potential therapy target in breast cancer; C2ORF40 gene product or C2ORF40-derived peptide might potentially be a suitable biotherapeutic reagent for breast and other types of tumors. Restoring C2ORF40 expression in tumors, either by epigenetic therapy or application of recombinant C2ORF40-derived peptide, may represent a promising therapeutic approach for breast cancers.
Materials and methods
Cell lines and breast tumor samples
All breast cancer cell lines used in this study were obtained from ATCC and were propagated in the media according to the conditions recommended by ATCC at 37°C in a 5% CO2 atmosphere. The human primary breast tumors were collected at the time of surgical resection at Hospital Universitario of Salamanca, Salamanca, Spain. The cancer tissues were snap-frozen down and storage at -80°C freezer. Collection and the use of patient samples were approved by the institutional ethics review board of the Hospital Universitario of Salamanca. Written informed consent was obtained from all patients for research using these tumor samples.
Transcriptional data sets used in study
We used three previously published transcriptional profiling data sets that contained both normal and breast cancer samples and four breast cancer data sets that included clinical and gene expression data (Table S1). The normal and primary human breast tumor samples in these data sets had been profiled with an Affymetrix microarray assay (either HG-U133B or HG U133 Plus 2.0) or Agilent oligo microarray (Table S1) since only HG-U133B, HG U133 Plus 2.0 and Agilent oligo microarrays contains the probes for C2ORF40. A lot of data sets were excluded in this study due to the lack of microarray probes for C2ORF40. The processed data from GEO website were downloaded for analysis.
Establishment of breast cancer cell line stably expressing or silencing C2ORF40
Human C2ORF40 cDNA was cloned into retroviral pBabe-puro plasmids to make overexpression retroviral vector. Two shRNAs against AACGAGAAGCACCTGTTCC and GACTAAAGTGGCCGTTGAT sequences of human C2ORF40 respectively were synthesized and cloned into pSuper vector according to instruction to generate C2ROF40 specific shRNA vectors (designated as shA and shB). Retroviral particles were produced as described previously.Citation27,Citation28 Briefly, C2ORF40 (or control empty) retroviral vectors along with packaging system pHit60 and pVSVG vectors were co-transfected into the HEK 293 Phoenix ampho packaging cells (ATCC). The virus containing supernatant was filtered through a 0.22 μm syringe filter. Retroviral transduction was performed by adding filtered supernatant to cultured cell lines in the presence of 2 μg/ml of polybrene (Sigma-Aldrich) and transduced cell lines were selected with 2 μg/ml puromycin. The expression level of C2ORF40 was confirmed by RT-PCR and western blotting analysis.
RT-PCR and quantitative Real-time RT-PCR
Cells at 50% to 70% confluence were trypsinized and total RNA was extracted with TRIzol Reagent (Invitrogen Life Technologies). One μg of total RNA was reverse transcribed using a First Strand Synthesis kit (Fermentas). RT-PCR and quantitative real-time RT-PCR (qRT-PCR) were performed to measure the expression of C2ORF40 and UBE2C. The primers designed for C2ORF40 were 5′-GGTACCAGCAGTTTCTCTACATG-3′ as forward and 5′-CAGCGTGTGGCAAGTCATGGTTAGT-3′ as reverse. The primers for UBE2C were 5′-TGATGTCTGGCGATAAAGGG-3′ as forward and 5′-TGATAGCAGGGCGTG AGGAA-3′ as reverse. GAPDH gene was used as an internal control using the forward primer 5′-GCCGCATCTTCTTTTGCGTCGC-3′ and reverse primer 5′-TCCCGTTCTCAGCCTTGACGGT-3′. RT-PCR was performed for 30 cycles. Each of the PCR products was directly loaded onto 2% agarose gel containing ethidium bromide and visualized under UV transilluminator. qRT-PCR was performed for 40 cycles and each PCR cycle including denaturing at 94°C for 30s, annealing for 30s at 54°C and extension at 72°C for 45s. PCR reactions were performed in triplicate for each sample and experiments were repeated a minimum of three times. Ct values were normalized against GAPDH RNA (ΔCt = Ct of C2ORF40/UBE2C - Ct of GAPDH). The relative C2ORF40 or UBE2C expression was calculated by 2^ (-ΔΔCt), where ΔΔCt = (ΔCt of sample) – (average ΔCt of three normal controls).
Western immunoblots
Cells in culture at 50% to 70% confluence were washed in ice-cold phosphate buffered saline (PBS) and lysed in RIPA lysis buffer (containing 25 mM TRIS-HCl, pH 7.6, 5 mM Hepes, 150 mM NaCl, 1% Nonidet-P40, 1% sodium deoxycholate, 0.1% SDS) supplemented with 1 x protease inhibitor cocktail (Roche Molecular Systems). Protein concentrations were determined using the Bio-Rad BCA protein assay kit (Bio-Rad Laboratories). For Western immunoblots, 30 μg of protein extracts per lane were electrophoresed with denaturing SDS-polyacrylamide gels (12%), transferred to PVDF membranes (Millipore). The membrane were blocked in TBST/5% skim milk for 1 h at room temperature, incubated with C2ORF40 (Santa Cruz Biotechnology) or UBE2C (Sigma-Aldrich) antibody at 1:500 dilution and then washed three times with TBST followed by incubation with HRP-conjugated secondary antibody. The signal was visualized using ECL detection reagent (Millipore).
DNA Extraction, sodium bisulfite modification and methylation analysis
Genomic DNA was extracted by standard SDS/proteinase K digestion followed by phenol-chloroform extraction and ethanol precipitation. Genomic DNA was converted with the EZ DNA Methylation Direct Kit (Zymo Research). Sodium bisulfite-treated DNA was amplified by PCR for region of -420 to +220 of C2ORF40 gene using four sets of primers as follows: 5′-GGTTTTGGAGTTTAGGGGT-3′, 5′- ACCCCTTAACCCTACCTAA-3′, 5′-AAATCCTCCCTCTAAATAACCA-3′ and 5′- GGGTTTTAGTATAGGAGTAGGA-3′. The PCR products were then sequenced.
Methylation-specific polymerase chain reaction (MSPCR) analysis was used for qualitative analysis of methylation. PCR amplification of the bisulfate-treated genomic DNAs was performed in two separate reactions with primer pairs specific for either the unmethylated (U) or the methylated (M) sequence. MSPCR primers are: M-forward primer: 5′-AGAGGATTTCGGTGGTATTCGTTC-3′; M-reverse primer: 5′-GACCGCGAATTATCCCTACG-3′. U-forward primer: 5′- GAGAGAGGATTTTGGTGGTATTTGTTTG-3′; M-reverse primer: 5′- AACAAACAAACACAACCACAAATTATCCCTACA-3′. If the sample only had unmethylated PCR band, it was called unmethylated. If the sample only had methylated PCR band, it was called methylation [denoted as methylation (++)]. If the sample had both unmethylated and methylated PCR band, it was called partial methylation [denoted as methylation (+)].
5-aza-2’-deoxycytidine treatment
Breast cancer cells were cultured in medium supplemented with 5-aza-2’-deoxycytidine (5-aza-dC, Sigma-Aldrich) at a concentration of 15 μM for three days and then subjected to RNA or genomic DNA extraction as described previously.
Cell proliferation and cologenic assay
Cell proliferation was measured by thiazolyl blue tetrazolium bromide (MTT) assay. Breast cancer cells transduced with C2ORF40 containing and control retroviruses were plated in 96-well cell culture plates (2 × 103 cells per well). Approximately 72 h later, MTT reagent was added to each well at 5 mg/ml in 10 μl and incubated for another 4 h at 37°C. 150 μl of DMSO was added to each well and mix vigorously to solubilize colored crystals produced within the living cells. Optical density was measured at 490 nm for the absorbance values. Cologenic assay was performed as described.Citation29 Cells transduced with C2ORF40 or control retroviruses were plated in 60-mm dishes. After incubation at 37°C for 15 d, formed colonies were fixed in methanol and stained with Giemsa solution (Sigma-Aldrich). A cluster of a minimum of 50 cells is considered a colony.
Wound healing and invasion assay
Transduced cells at 100% confluency were mechanically scratched using a 200-μL pipette tip to create the wound. Scratched cells were washed with PBS to remove the debris, and fresh culture media were added to allow wound healing. Phase-contrast images of the wound were taken at 0 and 48 h after the scratch to examine the cell migration into wounded areas. In invasion assay, transduced cells were seeded into 24-well Matrigel invasion chambers at 1 × 105 cells per well in triplicate (BD Biosciences). Inserts were placed into falcon companion plates containing 10% fetal bovine serum and incubated for 16 h for cell invasion. After the incubation, non-migrated cells in the top chamber were removed with a cotton swab. Cells on the underside of the membranes were fixed with paraformaldehyde, stained with 2.5% crystal violet, washed with PBS and photographed under the microscope. Numbers of migrated cells on the underside side in five random fields from each well were counted under microscope.
Flow cytometric analysis of cell cycle
For cell cycle assay,Citation28,Citation29 cells at proliferative log phase were trypsinized and rinsed twice with ice-cold PBS solution, then fixed by 75% ice-cold ethanol. The fixed cells were washed with ice-cold PBS and incubated at 37°C for 30 min in 1 ml of PBS solution containing 20 μg/ml RNase A (Fermentas) and stained with 20 μg/ml of propidium iodide (Sigma-Aldrich) at room temperature for 10 min. DNA content was then determined by flow cytometric analysis. The percentages of cells in G0/G1, S and G2/M phases were determined on BD FACS Calibur (Becton Dickinson) and data were analyzed with FlowJo software (Tree Star, Inc.).
Functional annotation and pathways analysis
A list of genes were defined to be significantly correlated (Pearson Correlation) with the mRNA expression levels of C2ORF40 by the criteria (absolute correlation coefficient ≤ 0.3 and FDR < 0.01 in all three data sets (GSE1456, GSE3494 and GSE6532), which were profiled by same platform of Affymetrix microarray. Pathways were identified with Ingenuity Pathway Analysis (IPA). The enrichment of specific functional groups (Gene Ontology (GO) terms) was analyzed by The Database for Annotation, Visualization and Integrated Discovery (DAVID) (http://david.abcc.ncifcrf.gov/).
Statistical analysis
The difference in C2ORF40 mRNA expression levels between normal and breast cancers was analyzed by Mann-Whitney U (two groups) or Kruskal-Wallis (three groups) test using publicly available data sets (Table S1). Spearman’s correlation coefficient and test were used to examine the relationship between C2ORF40 and UBE2C mRNA levels. Kaplan-Meier plots were constructed and a long rank test was used to determine differences among disease free and distant metastasis free survival curves according to C2ORF40 expression levels using publicly available data sets (Table S1). In addition, the relation between C2ORF40 expression and survival was explored in microarray data sets by dividing the cases from each cohort into a group with high (top one-third), moderate (middle one-third) and low (bottom one-third) level of expression. All analyses were performed by SPSS 11.5.0 for Windows. A two-tailed P value of less than 0.05 was considered to indicate statistical significance.
Additional material
Download Zip (411.8 KB)Acknowledgments
This work was supported by National Natural Science Foundation of China (grant numbers 81172528, 31271461 to G. W.); Doctoral Fund of Ministry of Education of China (grant number 20110131110035 to G. W.); Shandong Provincial Natural Science Foundation, China (grant number ZR2011HM034 to G. W.); by the National Institutes of Health, National Cancer Institute grant (grant number R01 CA116481 to J. H. M.); the Low Dose Scientific Focus Area, Office of Biological and Environmental Research, US Department of Energy (grant number DE-AC02–05CH11231 to J. H. M.); and Laboratory Directed Research and Development Program (LDRD) (to J. H.M.).
Submitted
01/17/2013
Revised
03/28/2013
Accepted
04/09/2013
Disclosure of Potential Conflicts of Interest
No potential conflict of interest was disclosed.
References
- Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin 2011; 61:69 - 90; http://dx.doi.org/10.3322/caac.20107; PMID: 21296855
- Zhang B, Beeghly-Fadiel A, Long J, Zheng W. Genetic variants associated with breast-cancer risk: comprehensive research synopsis, meta-analysis, and epidemiological evidence. Lancet Oncol 2011; 12:477 - 88; http://dx.doi.org/10.1016/S1470-2045(11)70076-6; PMID: 21514219
- Su T, Liu H, Lu S. [Cloning and identification of cDNA fragments related to human esophageal cancer]. Zhonghua Zhong Liu Za Zhi 1998; 20:254 - 7; PMID: 10920976
- Steck E, Breit S, Breusch SJ, Axt M, Richter W. Enhanced expression of the human chitinase 3-like 2 gene (YKL-39) but not chitinase 3-like 1 gene (YKL-40) in osteoarthritic cartilage. Biochem Biophys Res Commun 2002; 299:109 - 15; http://dx.doi.org/10.1016/S0006-291X(02)02585-8; PMID: 12435396
- Mirabeau O, Perlas E, Severini C, Audero E, Gascuel O, Possenti R, et al. Identification of novel peptide hormones in the human proteome by hidden Markov model screening. Genome Res 2007; 17:320 - 7; http://dx.doi.org/10.1101/gr.5755407; PMID: 17284679
- Yue CM, Deng DJ, Bi MX, Guo LP, Lu SH. Expression of ECRG4, a novel esophageal cancer-related gene, downregulated by CpG island hypermethylation in human esophageal squamous cell carcinoma. World J Gastroenterol 2003; 9:1174 - 8; PMID: 12800218
- Götze S, Feldhaus V, Traska T, Wolter M, Reifenberger G, Tannapfel A, et al. ECRG4 is a candidate tumor suppressor gene frequently hypermethylated in colorectal carcinoma and glioma. BMC Cancer 2009; 9:447; http://dx.doi.org/10.1186/1471-2407-9-447; PMID: 20017917
- Mori Y, Ishiguro H, Kuwabara Y, Kimura M, Mitsui A, Kurehara H, et al. Expression of ECRG4 is an independent prognostic factor for poor survival in patients with esophageal squamous cell carcinoma. Oncol Rep 2007; 18:981 - 5; PMID: 17786363
- Li LW, Yu XY, Yang Y, Zhang CP, Guo LP, Lu SH. Expression of esophageal cancer related gene 4 (ECRG4), a novel tumor suppressor gene, in esophageal cancer and its inhibitory effect on the tumor growth in vitro and in vivo. Int J Cancer 2009; 125:1505 - 13; http://dx.doi.org/10.1002/ijc.24513; PMID: 19521989
- Li W, Liu X, Zhang B, Qi D, Zhang L, Jin Y, et al. Overexpression of candidate tumor suppressor ECRG4 inhibits glioma proliferation and invasion. J Exp Clin Cancer Res 2010; 29:89; http://dx.doi.org/10.1186/1756-9966-29-89; PMID: 20598162
- Li LW, Yang Y, Li XY, Guo LP, Zhou Y, Lu SX. [Tumor-suppressing function of human esophageal cancer related gene 4 in esophageal squamous cell carcinoma]. Zhonghua Yi Xue Za Zhi 2010; 90:2713 - 7; PMID: 21162904
- Sabatier R, Finetti P, Adelaide J, Guille A, Borg JP, Chaffanet M, et al. Down-regulation of ECRG4, a candidate tumor suppressor gene, in human breast cancer. PLoS One 2011; 6:e27656; http://dx.doi.org/10.1371/journal.pone.0027656; PMID: 22110708
- Chin K, DeVries S, Fridlyand J, Spellman PT, Roydasgupta R, Kuo WL, et al. Genomic and transcriptional aberrations linked to breast cancer pathophysiologies. Cancer Cell 2006; 10:529 - 41; http://dx.doi.org/10.1016/j.ccr.2006.10.009; PMID: 17157792
- Neve RM, Chin K, Fridlyand J, Yeh J, Baehner FL, Fevr T, et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell 2006; 10:515 - 27; http://dx.doi.org/10.1016/j.ccr.2006.10.008; PMID: 17157791
- Stingl J, Caldas C. Molecular heterogeneity of breast carcinomas and the cancer stem cell hypothesis. Nat Rev Cancer 2007; 7:791 - 9; http://dx.doi.org/10.1038/nrc2212; PMID: 17851544
- Vanaja DK, Ehrich M, Van den Boom D, Cheville JC, Karnes RJ, Tindall DJ, et al. Hypermethylation of genes for diagnosis and risk stratification of prostate cancer. Cancer Invest 2009; 27:549 - 60; http://dx.doi.org/10.1080/07357900802620794; PMID: 19229700
- Esteller M. Dormant hypermethylated tumour suppressor genes: questions and answers. J Pathol 2005; 205:172 - 80; http://dx.doi.org/10.1002/path.1707; PMID: 15643671
- Esteller M. Epigenetic gene silencing in cancer: the DNA hypermethylome. Hum Mol Genet 2007; 16:Spec No 1 R50 - 9; http://dx.doi.org/10.1093/hmg/ddm018; PMID: 17613547
- Greenman C, Stephens P, Smith R, Dalgliesh GL, Hunter C, Bignell G, et al. Patterns of somatic mutation in human cancer genomes. Nature 2007; 446:153 - 8; http://dx.doi.org/10.1038/nature05610; PMID: 17344846
- Li LW, Li YY, Li XY, Zhang CP, Zhou Y, Lu SH. A novel tumor suppressor gene ECRG4 interacts directly with TMPRSS11A (ECRG1) to inhibit cancer cell growth in esophageal carcinoma. BMC Cancer 2011; 11:52; http://dx.doi.org/10.1186/1471-2407-11-52; PMID: 21288367
- Reddy SK, Rape M, Margansky WA, Kirschner MW. Ubiquitination by the anaphase-promoting complex drives spindle checkpoint inactivation. Nature 2007; 446:921 - 5; http://dx.doi.org/10.1038/nature05734; PMID: 17443186
- Williamson A, Wickliffe KE, Mellone BG, Song L, Karpen GH, Rape M. Identification of a physiological E2 module for the human anaphase-promoting complex. Proc Natl Acad Sci U S A 2009; 106:18213 - 8; http://dx.doi.org/10.1073/pnas.0907887106; PMID: 19822757
- Wagner KW, Sapinoso LM, El-Rifai W, Frierson HF, Butz N, Mestan J, et al. Overexpression, genomic amplification and therapeutic potential of inhibiting the UbcH10 ubiquitin conjugase in human carcinomas of diverse anatomic origin. Oncogene 2004; 23:6621 - 9; http://dx.doi.org/10.1038/sj.onc.1207861; PMID: 15208666
- Ozawa A, Lick AN, Lindberg I. Processing of proaugurin is required to suppress proliferation of tumor cell lines. Mol Endocrinol 2011; 25:776 - 84; http://dx.doi.org/10.1210/me.2010-0389; PMID: 21436262
- Mirabeau O, Perlas E, Severini C, Audero E, Gascuel O, Possenti R, et al. Identification of novel peptide hormones in the human proteome by hidden Markov model screening. Genome Res 2007; 17:320 - 7; http://dx.doi.org/10.1101/gr.5755407; PMID: 17284679
- Dang X, Podvin S, Coimbra R, Eliceiri B, Baird A. Cell-specific processing and release of the hormone-like precursor and candidate tumor suppressor gene product, Ecrg4. Cell Tissue Res 2012; 348:505 - 14; http://dx.doi.org/10.1007/s00441-012-1396-6; PMID: 22526622
- Hu Z, Huang G, Sadanandam A, Gu S, Lenburg ME, Pai M, et al. The expression level of HJURP has an independent prognostic impact and predicts the sensitivity to radiotherapy in breast cancer. Breast Cancer Res 2010; 12:R18; http://dx.doi.org/10.1186/bcr2487; PMID: 20211017
- Jones L, Wei G, Sevcikova S, Phan V, Jain S, Shieh A, et al. Gain of MYC underlies recurrent trisomy of the MYC chromosome in acute promyelocytic leukemia. J Exp Med 2010; 207:2581 - 94; http://dx.doi.org/10.1084/jem.20091071; PMID: 21059853
- Wei G, Ku S, Ma GK, Saito S, Tang AA, Zhang J, et al. HIPK2 represses beta-catenin-mediated transcription, epidermal stem cell expansion, and skin tumorigenesis. Proc Natl Acad Sci U S A 2007; 104:13040 - 5; http://dx.doi.org/10.1073/pnas.0703213104; PMID: 17666529