Abstract
Reduced fetal growth associates with endothelial dysfunction and cardiovascular risk in both young and adult offspring and the nitric oxide (NO) system has been implicated in these effects. Epigenetic processes are likely to underlie such effects, but there is to date no evidence that endothelial dysfunction in early life results from epigenetic processes on key genes in the NO system, such as NOS3 (eNOS) and ARG2 (arginase-2). We determined basal DNA methylation status in NOS3 and ARG2 promoters, and DNA methyltransferase 1 (DNMT1) effect on eNOS and arginase-2 expression using human endothelial cells isolated from umbilical arteries (HUAEC) and veins (HUVEC) from control and intrauterine growth restricted (IUGR) fetuses. Compared with cells from control pregnancies, eNOS protein and mRNA levels were increased in HUAEC, but decreased in HUVEC, from IUGR, while arginase-2 levels were increased in IUGR-HUVEC. The NOS3 promoter showed a decrease in DNA methylation at CpG -352 in IUGR-HUAEC, and an increase in IUGR-HUVEC, when compared with control cells. Methylation in the hypoxia response element of the NOS3 promoter was increased in IUGR-HUAEC and decreased in HUVEC. Methylation in the AGR2 promoter in IUGR-HUVEC was decreased in a putative HRE, and without changes in IUGR-HUAEC. Silencing of DNMT1 expression normalized eNOS expression in IUGR endothelial cells, and restored the normal response to hypoxia in HUVEC, without effects on arginase-2. This data suggest that eNOS expression in IUGR-derived endothelial cells is programmed by altered DNA methylation, and can be reversed by transient silencing of the DNA methylation machinery.
Introduction
Epidemiologic studies in the late 1980s in the UK showed the correlation between reduced fetal growth (i.e., low birth weight) and increased risk of later cardiometabolic diseases.Citation1,Citation2 Subsequently, the association between environmental cues during embryonic and fetal development, and risk of non-communicable diseases, such as obesity, diabetes and hypertension,Citation3 has been widely confirmed. Studies using animal models have established the detrimental effects of maternal malnutrition, placental dysfunction, and other factors on early embryonic and fetal development and on health consequences in later life.Citation4 Stimuli related to unhealthy maternal lifestyle and environment can induce physiological changes in the offspring that may represent adaptations to unfavorable intrauterine conditions and can also confer benefit in coping if such environmental conditions persist after birth.Citation5 Insights from developmental biology suggest that epigenetic mechanisms underlie such effects.Citation6
Reduced fetal growth is not only associated with adult vascular dysfunction. Umbilical and placental vessels derived from intrauterine growth restricted (IUGR) fetuses show impaired endothelium-dependent vasodilation compared with appropriately grown for gestational age fetuses,Citation7,Citation8 and this is also observed in the peripheral circulation of low birth weight neonates.Citation9 These functional changes associate to altered expression of several vascular mediators in fetoplacental-derived endothelial cells (EC), including eNOS and arginase-2.Citation8 Additionally, reduced arterial compliance and endothelial dysfunction correlated with IUGR are found in pre-pubertal,Citation10 adolescentCitation10 and young-adults,Citation11 suggesting that IUGR establishes a sustained alteration in vascular function, which may be driven by epigenetic mechanisms.Citation6
Conversely, epigenetic mechanisms play a key role in vascular development, physiology and pathophysiology. Basal expression of several genes crucial for endothelial function, such as eNOSCitation12-Citation14 and others,Citation15-Citation19 is controlled by DNA methylation and histone posttranslational modifications. These mechanisms operate in a gene-specific manner in EC, suggesting the existence of an endothelial-specific epigenetic code,Citation20 an idea convincingly demonstrated in NOS3 gene in normal endothelial and non-EC.Citation21 However, there is no data addressing whether epigenetic mechanisms regulate the expression of another critical protein for the l-arginine/NO pathway, such as arginase-2, involved in the development of endothelial dysfunction.Citation22
In the present study, we examined the role of DNMT1-dependent methylation, which preserves the DNA methylation pattern after mitotis,Citation23,Citation24 on the altered eNOS expression in cultured HUAEC and HUVEC from IUGR pregnancies. In order to determine whether DNMT1 was involved in the blunted response to the hypoxia-mediated eNOS downregulation observed in IUGR-HUVEC, we evaluated this response in cells subjected to DNMT1 knockdown. Finally, in order to determine whether these effects were specific to eNOS, we studied the expression of arginase-2, a crucial eNOS counteracting protein under both normal and pathological conditions,Citation22 and investigated whether its expression was similarly regulated by DNMT1.
Results
Basal expression of eNOS and arginase-2 in IUGR vs. control endothelial cells
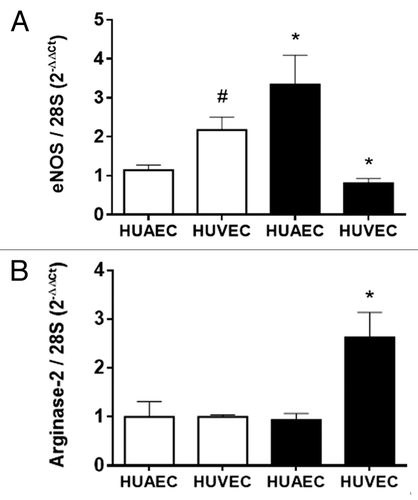
Endothelial NOS mRNA levels were increased in IUGR-HUAEC and decreased in IUGR-HUVEC compared with their corresponding control cells (). Conversely, arginase-2 mRNA levels were increased only in IUGR-HUVEC (). Furthermore, eNOS expression observed in IUGR-HUAEC was similar to that observed in control HUVEC, while IUGR-HUVEC exhibited an expression comparable to control HUAEC. These changes are in accordance to those reported for the protein levels of eNOS and arginase-2 in these cell types.Citation8,Citation25,Citation26
Figure 1. Expression of eNOS and arginase-2 in normal and IUGR cells relative to normal HUAEC. Quantification of eNOS (A) and arginase-2 (B) mRNA levels in control (open bars, n = 5) and IUGR (solid bars, n = 5) cultured HUAEC and HUVEC. Values are mean ± SEM *p < 0.05, vs. corresponding normal cells, #p < 0.05 vs. control HUAEC, one-way ANOVA.
DNA methylation status in NOS3 promoter of IUGR endothelial cells
We studied the methylation status of CpGs located in the NOS3 proximal promoter (-51 to -351), where its basal expression is controlled,Citation27 the shear stress response element (SSRE, -976 to -992)Citation28 and the hypoxia response element (HRE, -5369 to -5375),Citation29 which represent important stimuli controlling endothelial function (). Methylation status in the promoter region of the NOS3 gene was comparable in HUAEC, placental chorionic artery endothelial cells (PLAEC) and HUVEC isolated from control tissues (). However, there were significant changes in the level of methylation at three different CpGs within the promoter of NOS3, namely CpGs -352, -5369 and -5375 relative to the reported transcription start site. Methylation was reduced at CpG -352, but increased at CpGs -5369 and -5375 in control HUVEC, compared with HUAEC and PLAEC. In contrast, in IUGR-EC there was only a change in methylation at a single CpG located at at CpG -352, where methylation was higher in HUVEC compared with HUAEC ().
Figure 2. DNA methylation pattern in the NOS3 promoter of placental and umbilical endothelial cells. (A) Regions analyzed included the core promoter (-100 to -51), proximal promoter (-361 and -352), the shear stress response element (-992 to -976) and the hypoxia response element (-5375 and -5369). Specific CpG methylation (%) determined by pyrosequencing of bisulfite-treated DNA from HUAEC (white bars, n = 9), PLAEC (light gray bars, n = 4) and HUVEC (dark gray bars, n = 9) isolated from control (B) and IUGR (C) placentae. Values are mean ± SEM *p < 0.05, vs. HUAEC, two-way ANOVA.
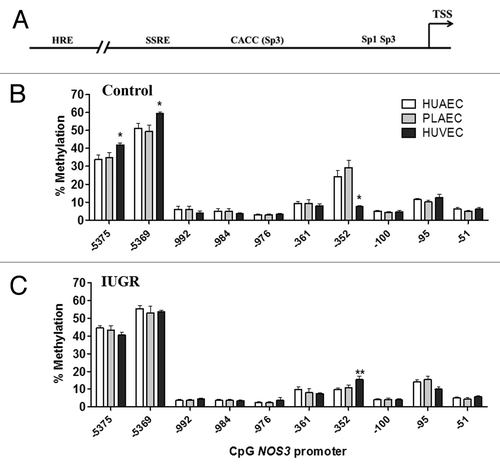
Interestingly, changes in the methylation status in IUGR-EC compared with control cells were limited to the three above-mentioned CpGs. In IUGR-HUAEC methylation at CpG -352 was decreased, while CpG -5375 showed an increased in the methylation level compared with control HUAEC (). Similar changes were found in IUGR-PLAEC, with a decrease at CpG -352 and increase at CpG -5375 compared with control PLAEC (). In contrast, in IUGR-HUVEC there was an increase in CpG -352, but a decrease in CpG -5369methylation compared with control HUVEC ().
Figure 3. Comparison of DNA methylation status in the NOS3 promoter of normal and IUGR cells. Specific CpG methylation (%) in normal and IUGR-HUAEC (A), PLAEC (B) and HUVEC (C) determined by pyrosequencing of bisulfite-treated DNA samples. Values are mean ± SEM ***p < 0.001, *p < 0.05, vs. corresponding control EC, two-way ANOVA.
DNA methylation status in ARG2 promoter of IUGR endothelial cells
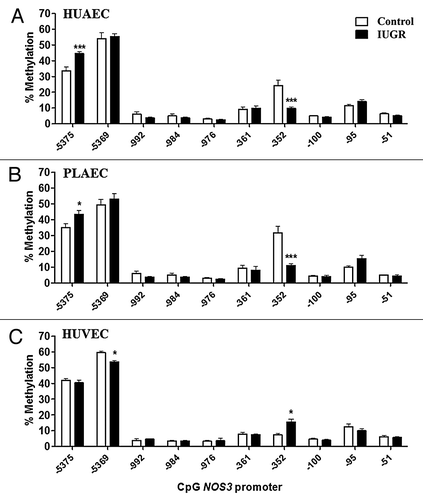
There is no report characterizing the control of ARG2 promoter activity in human EC. In this study, we analyzed the methylation status in the core and proximal ARG2 promoter, which presents a high number of CpGs within several putative transcription factor binding sites (). ARG2 promoter CpGs were numbered according to the reported ATG for this gene (). Methylation status in the promoter region of the ARG2 gene was comparable in HUAEC, PLAEC and HUVEC isolated from control placentae with a single difference between PLAEC and HUVEC at CpG -465 (). Similarly, CpG -465 was differentially methylated in IUGR-EC, being decreased in HUVEC compared with HUAEC ().
Figure 4. Prediction of transcription factors binding sites in ARG2 promoter. Cognate sequences for human transcription factors (underlined) and CpGs (highlighted in yellow) in the proximal and core promoter of ARG2 gene. Proposed transcription factor were obtained by in silico analysis of ARG2 promoter sequence with MatInspector software, and CpGs were numbered according to the position regarding the reported ATG.
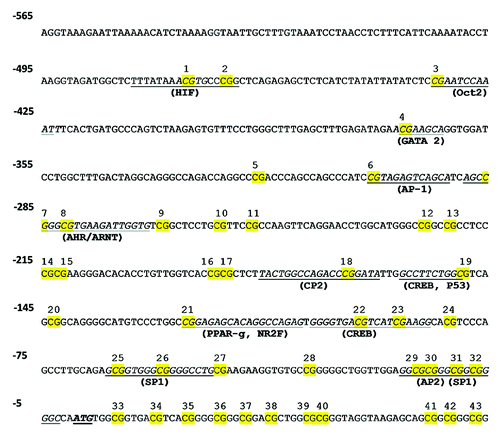
Figure 5. DNA methylation status in the ARG2 promoter of placental and umbilical endothelial cells. (A) Regions analyzed included the core (-33 to -7) and proximal (-471 and -281) promoter, where putative transcription factors or consensus binding sites are indicated. Specific CpG methylation (%) was determined by pyrosequencing of bisulfite-treated DNA from HUAEC (white bars, n = 9), PLAEC (light gray bars, n = 4) and HUVEC (dark gray bars, n = 9) isolated from normal (B) and IUGR (C) placentae. Values are mean ± SEM *p < 0.05 vs. HUAEC, #p < 0.05 vs. PLAEC, two-way ANOVA.
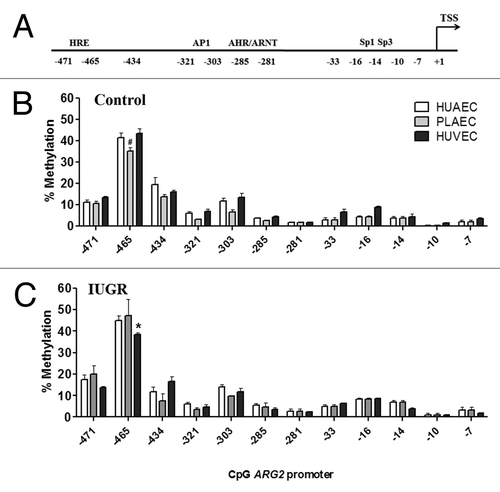
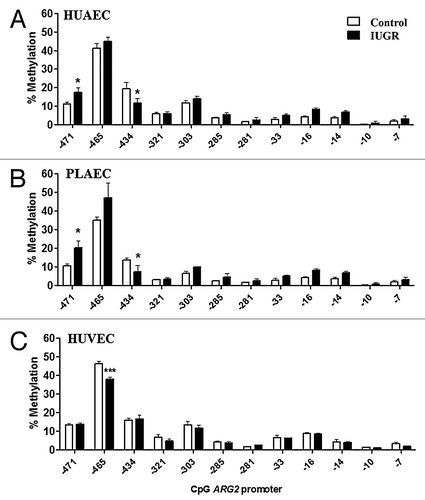
Changes in ARG2 promoter methylation status were comparable between IUGR-HUAEC and PLAEC (), similar to those observed for the NOS3 promoter (). Methylation at CpG -434 was reduced in IUGR-HUAEC () and PLAEC (), while it was increased at CpG -471 compared with the corresponding control cells. In IUGR-HUVEC there was a single change in CpG methylation status at -465 compared with control HUVEC ().
Figure 6. Comparison of DNA methylation status in the ARG2 promoter of normal and IUGR cells. Specific CpG methylation (%) in normal and IUGR-HUAEC (A), PLAEC (B) and HUVEC (C) determined by pyrosequencing of bisulfite treated DNA samples. Values are mean ± SEM ***p < 0.001, *p < 0.05, vs. corresponding control EC, two-way ANOVA.
Effect of mitotically heritable DNA methylation on basal eNOS and arginase-2 expression in IUGR-HUAEC and HUVEC
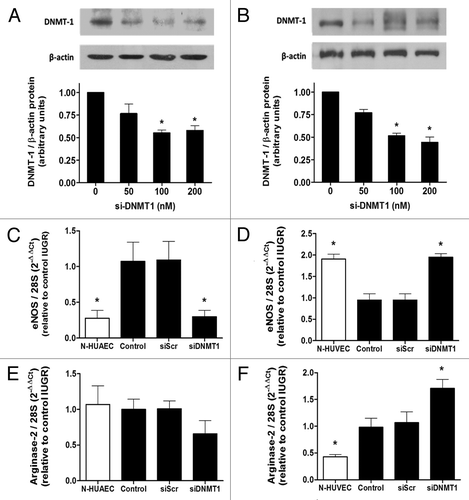
In order to determine the role of mitotically heritable DNA methylation on the basal expression of eNOS mRNA, DNA methyltransferase-1 (DNMT1) protein was silenced with a previously described specific siRNA (detailed in methods) in EC isolated from IUGR placentae. Treatment with the silencer (siDNMT1) partially reduced (~50%) the expression of DNMT1 protein in HUAEC () and HUVEC (). In IUGR-HUAEC, basal eNOS mRNA levels were not altered by the treatment with a non-specific silencing RNA (siScr); however, siDNMT1 reduced eNOS mRNA, restoring it to normal levels (). In contrast, in IUGR-HUVEC the reduced basal eNOS mRNA levels were not affected by siScr, but were increased after transient silencing of DNMT1 normalizing eNOS expression in this cell type (). In IUGR-HUAEC, the levels of arginase-2 mRNA were not affected either by siScr or siDNMT1 (). On the other hand, in IUGR-HUVEC, silencing of DNMT1 induced a further increase in arginase-2 mRNA levels, not seen with siScr ().
Figure 7. Effect of DNMT1 silencing on eNOS and arginase-2 mRNA levels in IUGR endothelial cells. Silencing of DNA methyltransferase-1 (DNMT-1) induced a concentration-dependent downregulation of this protein in HUAEC (A) and HUVEC (B). Quantification of eNOS and arginase-2 mRNA levels in IUGR-HUAEC (C and E, respectively) and IUGR-HUVEC (D and F, respectively) (solid bars, n = 5) in basal conditions (control), and cells treated with an non-specific siRNA (siScr) or specific siRNA against DNMT1 (siDNMT1). Levels of mRNA are expressed as 2-ΔΔCT referred to control condition and compared with basal levels in normal cells (open bars, n = 5). Values are mean ± SEM *p < 0.05, vs. control, one-way ANOVA.
Effect of DNMT1 silencing on the IUGR-HUVEC response to hypoxia
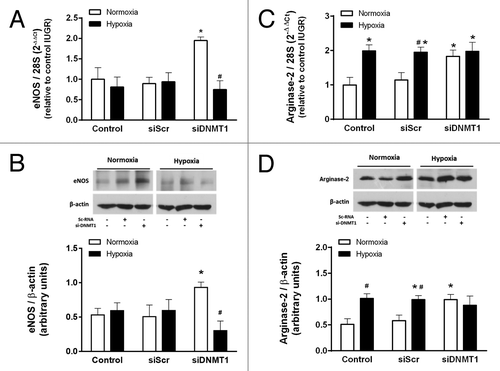
It is reported that IUGR-HUVEC present a hypoxia-like phenotype, characterized by absent downregulation of eNOS expression in response to low oxygen levels.Citation25 In order to determine whether this phenotype is influenced by DNA methylation, the response to 24 h of hypoxia was tested in IUGR-HUVEC in which DNMT1 was transiently silenced. The hypoxic response was evaluated in terms of eNOS and arginase-2 expression. In IUGR-HUVEC, eNOS mRNA levels were not affected in normoxia or hypoxia by the treatment with the siScr; however, siDNMT1 restored the normal downregulation of eNOS mRNA in response to hypoxia (). This effect was also observed at the protein level, as eNOS protein was decreased after the hypoxic challenge (). In contrast, in IUGR-HUVEC, levels of arginase-2 mRNA and protein increased in response to hypoxia, and this was not affected by siScr. siDNMT1 induced a basal increase in arginase-2 mRNA and protein, which were not further affected by hypoxia ().
Figure 8. Effect of DNMT1 silencing on the response to hypoxia in IUGR-HUVEC. Quantification of eNOS (mRNA, A; protein, B) and arginase-2 (mRNA, C; protein, D) levels in IUGR-HUVEC exposed to normoxia (5% oxygen, open bars, n = 5) or hypoxia (2% oxygen, solid bars, n = 5) in basal conditions (control), or in the presence of an unspecific siRNA (siScr) and a siRNA against DNA methyltransferase-1 (siDNMT1). mRNA levels are expressed as 2-ΔΔCT referred to control condition in normoxia. Values are mean ± SEM *p < 0.05 vs. control, #p < 0.05 vs. corresponding condition in normoxia, two-way ANOVA.
Discussion
This study demonstrates that changes in eNOS and arginase-2 proteins measured in cultured IUGR-HUAEC and HUVEC are paralleled by changes in mRNA levels, suggesting that IUGR modulates the transcriptional regulation of eNOS and arginase-2 in these cells. Notably, the changes in DNA methylation in the NOS3 promoter in IUGR EC occurred in the same CpGs that were differentially methylated between control arteries and vein. The DNA methylation pattern in the ARG2 promoter was comparable bewteen control endothelium, but different between arterial and venous EC in IUGR. Partial silencing of DNMT1 normalized eNOS mRNA levels in IUGR-HUAEC and HUVEC, but did not diminish the increased expression of arginase-2 in IUGR-HUVEC. Similarly, DNMT1 silencing recovered the normal response to hypoxia in terms of eNOS mRNA downregulation in IUGR-HUVEC. All together, these data showed that arginase-2 and eNOS expression are differentially regulated in IUGR-EC, and influenced by DNMT1 in a cell- and gene-specific manner.
Arginase activity is the main l-arginine consuming pathway in endothelial cell,Citation30 and a decreased eNOS to arginase activity ratio has been implicated in the development of vascular pathologies.Citation22 Recently, it was reported that eNOS and arginase-2 are differentially expressed in HUAEC and HUVEC derived from pregnancies with IUGR.Citation8 Endothelial NOS expression in IUGR-HUAEC has been reported to be increased,Citation8,Citation31 and similar results were found in IUGR-PLAEC,Citation32,Citation33 while eNOS expression in IUGR-HUVEC has been shown to be reduced in vivo and in vitro.Citation25,Citation34,Citation35 For arginase-2, only IUGR-HUVEC showed an increased mRNA expression, which relates with the reported protein levels.Citation8 The dissimilar changes in basal eNOS expression between IUGR-HUAEC and HUVEC found here apparently represent differences between vessel types (i.e., artery vs. vein), and could result from the altered blood flow and oxygen level characteristics of IUGR pregnancies.Citation36 It is widely believed that the maturation of blood vessels (i.e., blood vessel structure, identity and function) is influenced by hemodynamic factors and oxygen level.Citation37,Citation38 In fact, a brief exposure of near-term pregnant rats to hypoxia epigenetically programs an altered eNOS expression in pulmonary EC,Citation39 supporting the crucial role of oxygen in early endothelial programming. Interestingly, in IUGR-EC the altered arginase-2 and eNOS mRNA levels persist under standard culture conditions up to 4 weeks, suggesting that basal gene transcription is differentially controlled in IUGR compared with control cells.
Growing evidence reveals the presence of epigenetic markers in different cell types from individuals with altered fetal growth, indicating that these markers have a functional role.Citation3,Citation5 Findings in whole umbilical cords are mainly related to altered glucose metabolism and insulin resistance.Citation40,Citation41 However, considering that epigenetic mechanisms are cell specific, the significance of these altered epigenetic markers remains unknown. The present study is the first to show an epigenetic effect on eNOS expression in human EC derived from fetuses with altered growth trajectories. Although these results show the in vitro methylation status of NOS3 and ARG2 genes in a small number of human samples, which could be considered a limitation, these changes were analyzed in a specific cell type contributing to identify and clarify their potential long-term consequences on endothelial function.
Comparison of the NOS3 promoter DNA methylation pattern in normal EC showed differences at three specific CpGs between arterial and venous EC. It is worth noting that the differentially methylated CpGs at -5369 and -5375 correspond to the reported hypoxia response element.Citation29 Both of these CpGs showed a lower methylation in control PLAEC and HUAEC compared with HUVEC. Whether this participates in the differential regulation of eNOS expression in response to hypoxiaCitation26 remains to be determined. The other CpG that was differentially methylated between normal EC was located at -352 from the transcription start site, showing a higher methylation in HUAEC compared with HUVEC. There is only one previous report of the methylation status of the eNOS promoter in EC, showing that CpG -352 is differentially methylated between HUVEC and human dermal microvascular EC,Citation42 the latter having a methylation status comparable to that found in HUAEC and PLAEC in our study. These data suggest that CpG -352 might play a role in the differential regulation of basal eNOS expression in arterial and venous endothelium.
On the other hand, comparison of NOS3 promoter DNA methylation between control and IUGR EC showed that changes in the latter group were restricted to those CpGs that were differentially methylated in control EC, especially CpG -352. Considering that eNOS protein level is higher in normal HUVEC than HUAEC,Citation26 these results could explain the basal eNOS expression in IUGR cells which is increased in HUAEC, and decreased in HUVEC, reinforcing the proposed importance of CpG -352 in the regulation of basal eNOS expression. Moreover, most of the differences in NOS3 promoter DNA methylation observed between control EC were reduced between IUGR cells, suggesting that IUGR restricts the epigenetic diversity in EC.
To our knowledge there are no previous studies of the methylation of the ARG2 promoter in EC. Contrary to the NOS3 promoter, the comparison of ARG2 promoter between control EC showed only one CpG (-465) differentially methylated between PLAEC and HUVEC. Changes in DNA methylation between control and IUGR cells presented a similar pattern, where PLAEC and HUAEC showed comparable changes. However, the lack of reports showing functional significance of the sites with changes in DNA methylation status in this promoter does not clarify whether or not basal arginase-2 expression is influenced importantly by DNA methylation. This may agree with the idea that epigenetic mechanisms in EC mainly control the expression of crucial cell-specific genes,Citation20,Citation21 which perhaps does not include arginase-2.
Considering that altered eNOS expression may result from a mitotically-inherited epigenetic program, DNMT1 was transiently silenced in IUGR cell. DNMT1 knockdown restored eNOS mRNA levels in IUGR-HUAEC and HUVEC to control values. However, siDNMT1 did not normalize arginase-2 expression but further increased its expression in IUGR-HUVEC, without any effect in IUGR-HUAEC. This differential response to DNA methylation inhibition has been reported for several genes in cell lines, where DNMT1 has been silenced or inhibited.Citation43,Citation44 Moreover, eNOS expression in normal HUVEC is not altered by DNMT inhibition, but is importantly increased in vascular smooth muscle cells, and this induction is several times higher in HeLa cells.Citation42 Similarly, siDNMT1 had a differential effect in HUAEC and HUVEC, reducing and increasing basal eNOS expression, respectively. These different responses may result from the interaction of DNMTs with other chromatin modifying enzymes that normally operate in the NOS3 promoter in response to changes in oxygen levels.Citation45 It is worth noting that the effect of DNMT inhibition on gene expression could also result from altered expression of regulatory mechanisms, such as transcription factors and chromatin remodelers, which will finally modify the expression of eNOS. Independently of the precise mechanisms operating in our model, we hypothesize that after DNMT1 silencing in normoxia these mechanisms would be recruited establishing a cell-specific expression pattern.
Chronic hypoxia is an important issue en IUGR pregnancies.Citation36 HUVEC isolated from IUGR pregnancies show reduced eNOS expression in vitro compared with HUVEC from control pregnancies,Citation25 a pattern similar to that observed in control HUVEC under hypoxia.Citation25,Citation45,Citation46 Thus, in addition, epigenetic processes may affect such responses to hypoxia, which may have further clinical significance.Citation47 DNMT1 knockdown restored the hypoxic-induced downregulation of eNOS in IUGR-HUVEC at both mRNA and protein levels. Here, we showed that the IUGR-HUVEC have a persistent hypoxia-like phenotype and absent responses to normoxia in terms of eNOS expression,Citation25 which can be restored by DNMT1 silencing. These data suggest that DNA methylation-mediated control of eNOS gene promoter in IUGR cells controls not only basal expression but also cellular responses to stressors.
In summary, this study showed, for the first time, in a limited number of human samples, that altered in vitro expression of eNOS and arginase-2 in IUGR-derived EC is influenced by epigenetic mechanisms, supporting the notion that altered fetal growth is accompanied by altered epigenetic effects on endothelial function. Furthermore, these epigenetic changes can limit the vascular response to stressors such as hypoxia. Whether these alterations have an impact on long-term cardiovascular functions merits further examination.
Methods
Ethics statement and samples collection
The protocol of this study was performed following the ethical approval from the “Comité de Ética en Investigación,” Faculty of Medicine, Pontificia Universidad Católica de Chile, according with the principles outlined in the Declaration of Helsinki for the use of human tissue. Patients provided a written informed consent previous to the obtention of their placenta and umbilical cord samples. Placentae were collected after delivery from full-term normal (control group) and IUGR-diagnosed pregnancies from normotensive, non-smoking, non-alcohol or drug consuming mothers, without intrauterine infection or any other medical or obstetrical complication. Gestational age estimated by ultrasonography before the 12th week of pregnancy was considered; IUGR was defined as fetal body weight below the tenth centile for gestational age and sex as well as a low abdominal circumference according to standard growth chart as reported for Chilean population.Citation48
Both, maternal and newborn characteristics of compared groups are described in . Maternal age, parity, gestational age at birth and newborn gender were not different between subjects from normal or IUGR groups. Patients from the IUGR group had a lower mean birth weight, size and ponderal index compared with normal group. All IUGR new born were ranged between percentile 2 and 7, confirming the IUGR diagnose, while the new born from normal group ranged between percentile 25 and 90. Increased frequency of cesarean deliveries in the IUGR group resulted mainly from fetal non-reassuring state based on antepartum detection of positive contraction stress test, or oligohydramnios along with altered fetal umbilical Doppler test. In contrast in the normal group, C-section occurred after failed labor induction.
Table 1. Maternal and newborn group characteristics
Cell culture
Human endothelial cells from umbilical artery (HUAEC), umbilical vein (HUVEC) and placental chorionic plate arteries (PLAEC) were isolated by collagenase digestion from placentae and umbilical cord of control and intrauterine growth restricted (IUGR) fetuses. Cell were cultured in medium 199, and exposed for 24 h to normoxia (5% O2) or hypoxia (2% O2) prior to extraction of proteins, mRNA or DNA. Oxygen levels for normoxia and hypoxia were set based on those reported for normal and IUGR pregnancies, respectively (discussed in Krause et al., 2012).Citation26
Western Blotting
Proteins from HUAEC and HUVEC (70 μg) were separated by SDS-PAGE, blotted and probed with antibodies against eNOS, arginase-2 and β-actin. Proteins were detected by enhanced chemiluminescence and quantified by densitometry using Image J (NIH) and values were expressed as arbitrary units.
Quantitative PCR
Polymerase chain reactions were performed using oligonucleotide primers for human eNOS and arginase-2. The expression of the ribosomal subunit 28S was used as internal reference. Oligonucleotide primers were: 28S (sense) 5′-TTG AAA ATC CGG GGG AGA G-3′, 28S (antisense) 5′-ACA TTG TTC CAA CAT GCC AG-3′; arginase 2 (sense) 5′- CCA TCC TGA AGA AAT CCG TC-3′, arginase 2 (antisense) 5′- CTA ATG GTA CCG ATT GCC AG-3′; eNOS (sense) 5′-CCA GCT AGC CAA AGT CAC CAT-3′, eNOS (antisense) 5′-GTC TCG GAG CCA TAC AGG ATT-3′. Quantification was performed using the 2-ΔΔCT method, where –ΔΔCT(eNOS)X = - (ΔCT (eNOS/28S) X - ΔCT (eNOS/28S) reference sample), and –ΔΔCT(arginase-2)X = - (ΔCT (arginase-2/28S) X - ΔCT (arginase-2/28S) reference sample). Being CT the threshold cycle for the target sequence (arginase-2, eNOS), ΔCT the difference between the target sequence and a reference sequence (28S), “X” the analyzed sample (IUGR or normal), and the “reference sample” the control group. The results were expressed as arbitrary units regarding the reference group.
DNA methylation
Methylation status of the promoter regions of NOS3 and ARG2 genes was determined using bisulfite modification coupled to DNA sequencing.Citation49 Briefly, total DNA extracts from EC were treated with sodium bisulphite to convert unmethylated cytosine to uracil, and promoter regions were amplified by PCR using specific primers for NOS3 and ARG2 gene promoter (). One of the PCR primers was designed including a 5′-biotin tag, which allowed the purification of the amplicon directly from the PCR mix. Then biotin-containing amplified DNA strand was transferred to a PyroMark Q96 MD pyrosequencer (Qiagen) for sequencing at Southampton University (UK). Site-specific CpG methylations were determined as percentage, comparing the signal intensity of non-bisulfite-sensitive CpG and bisulfite-sensitive CpG.
Table 2. Primers for amplification and pyrosequencing of bisulfite-treated DNA samples
Transcription factors binding site prediction
Prediction of binding sites for transcription factors in ARG2 promoter was performed with the online software MatInspector,Citation50 selected transcription factors were chosen considering a cut-off of 0.900 for matrix similarity.
DNMT-1 knockdown
Both DNMT-1 (sc-35204) and control (sc-37007) siRNAs was purchased from Santa Cruz Biotechnology, and transfection was performed according to the protocol proposed by the manufacturer as well as has it been reported.Citation51 Briefly, sub-confluent IUGR cells (30–50%) were transfected with 100 nM of siRNA using Oligofectamine transfection reagent (Invitrogen) in Opti-MEM reduced serum medium (Invitrogen) for 6 h. The medium was removed and replaced with fresh DMEM supplemented with 10% fetal calf serum. Cells were grown in “normoxia” up to one passage further, and harvested upon confluence after being exposed to the different conditions proposed.
Statistical Analysis
Comparisons between two or more groups were performed by means of Student’s unpaired t-test and analysis of variance (ANOVA), respectively. All the analyses were performed with the statistical software Graphpad Prism 5.03 (GraphPad Software Inc.). p < 0.05 was considered the cut-off for statistical significance.
| Abbreviations: | ||
| IUGR | = | intrauterine growth restriction |
| DNMT1 | = | DNA methyltransferase 1 |
| EC | = | endothelial cells |
| eNOS | = | endothelial nitric oxide synthase |
| HUAEC | = | human umbilical artery endothelial cells |
| PLAEC | = | placental chorionic artery endothelial cells |
| HUVEC | = | human umbilical vein endothelial cells |
Acknowledgments
MAH is supported by the British Heart Foundation. Fondecyt 1120928 & 1130801 and Becas Chile (Pasantía Doctoral). EMU holds a CONICYT-Chile fellowship.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Barker DJ, Osmond C. Infant mortality, childhood nutrition, and ischaemic heart disease in England and Wales. Lancet 1986; 1:1077 - 81; http://dx.doi.org/10.1016/S0140-6736(86)91340-1; PMID: 2871345
- Barker DJ, Hales CN, Fall CH, Osmond C, Phipps K, Clark PM. Type 2 (non-insulin-dependent) diabetes mellitus, hypertension and hyperlipidaemia (syndrome X): relation to reduced fetal growth. Diabetologia 1993; 36:62 - 7; http://dx.doi.org/10.1007/BF00399095; PMID: 8436255
- Hanson M, Godfrey KM, Lillycrop KA, Burdge GC, Gluckman PD. Developmental plasticity and developmental origins of non-communicable disease: theoretical considerations and epigenetic mechanisms. Prog Biophys Mol Biol 2011; 106:272 - 80; http://dx.doi.org/10.1016/j.pbiomolbio.2010.12.008; PMID: 21219925
- McMillen IC, Robinson JS. Developmental origins of the metabolic syndrome: prediction, plasticity, and programming. Physiol Rev 2005; 85:571 - 633; http://dx.doi.org/10.1152/physrev.00053.2003; PMID: 15788706
- Gluckman PD, Hanson MA, Buklijas T, Low FM, Beedle AS. Epigenetic mechanisms that underpin metabolic and cardiovascular diseases. Nat Rev Endocrinol 2009; 5:401 - 8; http://dx.doi.org/10.1038/nrendo.2009.102; PMID: 19488075
- Krause B, Sobrevia L, Casanello P. Epigenetics: new concepts of old phenomena in vascular physiology. Curr Vasc Pharmacol 2009; 7:513 - 20; http://dx.doi.org/10.2174/157016109789043883; PMID: 19485890
- Dong YL, Green KE, Vegiragu S, Hankins GD, Martin E, Chauhan M, et al. Evidence for decreased calcitonin gene-related peptide (CGRP) receptors and compromised responsiveness to CGRP of fetoplacental vessels in preeclamptic pregnancies. J Clin Endocrinol Metab 2005; 90:2336 - 43; http://dx.doi.org/10.1210/jc.2004-1481; PMID: 15623815
- Krause BJ, Carrasco-Wong I, Caniuguir A, Carvajal J, Farías M, Casanello P. Endothelial eNOS/arginase imbalance contributes to vascular dysfunction in IUGR umbilical and placental vessels. Placenta 2013; 34:20 - 8; http://dx.doi.org/10.1016/j.placenta.2012.09.015; PMID: 23122700
- Martin H, Gazelius B, Norman M. Impaired acetylcholine-induced vascular relaxation in low birth weight infants: implications for adult hypertension?. Pediatr Res 2000; 47:457 - 62; http://dx.doi.org/10.1203/00006450-200004000-00008; PMID: 10759151
- Lurbe E, Torro MI, Carvajal E, Alvarez V, Redón J. Birth weight impacts on wave reflections in children and adolescents. Hypertension 2003; 41:646 - 50; http://dx.doi.org/10.1161/01.HYP.0000048341.52293.7C; PMID: 12623973
- Oren A, Vos LE, Bos WJ, Safar ME, Uiterwaal CS, Gorissen WH, et al. Gestational age and birth weight in relation to aortic stiffness in healthy young adults: two separate mechanisms?. Am J Hypertens 2003; 16:76 - 9; http://dx.doi.org/10.1016/S0895-7061(02)03151-5; PMID: 12517688
- Fish JE, Matouk CC, Rachlis A, Lin S, Tai SC, D’Abreo C, et al. The expression of endothelial nitric-oxide synthase is controlled by a cell-specific histone code. J Biol Chem 2005; 280:24824 - 38; http://dx.doi.org/10.1074/jbc.M502115200; PMID: 15870070
- Gan Y, Shen YH, Wang J, Wang X, Utama B, Wang J, et al. Role of histone deacetylation in cell-specific expression of endothelial nitric-oxide synthase. J Biol Chem 2005; 280:16467 - 75; http://dx.doi.org/10.1074/jbc.M412960200; PMID: 15722551
- Rössig L, Urbich C, Brühl T, Dernbach E, Heeschen C, Chavakis E, et al. Histone deacetylase activity is essential for the expression of HoxA9 and for endothelial commitment of progenitor cells. J Exp Med 2005; 201:1825 - 35; http://dx.doi.org/10.1084/jem.20042097; PMID: 15928198
- Peng Y, Jahroudi N. The NFY transcription factor inhibits von Willebrand factor promoter activation in non-endothelial cells through recruitment of histone deacetylases. J Biol Chem 2003; 278:8385 - 94; http://dx.doi.org/10.1074/jbc.M213156200; PMID: 12511565
- Wu J, Iwata F, Grass JA, Osborne CS, Elnitski L, Fraser P, et al. Molecular determinants of NOTCH4 transcription in vascular endothelium. Mol Cell Biol 2005; 25:1458 - 74; http://dx.doi.org/10.1128/MCB.25.4.1458-1474.2005; PMID: 15684396
- Dutta D, Ray S, Vivian JL, Paul S. Activation of the VEGFR1 chromatin domain: an angiogenic signal-ETS1/HIF-2alpha regulatory axis. J Biol Chem 2008; 283:25404 - 13; http://dx.doi.org/10.1074/jbc.M804349200; PMID: 18625704
- Lagarkova MA, Volchkov PY, Philonenko ES, Kiselev SL. Efficient differentiation of hESCs into endothelial cells in vitro is secured by epigenetic changes. Cell Cycle 2008; 7:2929 - 35; http://dx.doi.org/10.4161/cc.7.18.6700; PMID: 18814342
- Kanki Y, Kohro T, Jiang S, Tsutsumi S, Mimura I, Suehiro J, et al. Epigenetically coordinated GATA2 binding is necessary for endothelium-specific endomucin expression. EMBO J 2011; 30:2582 - 95; http://dx.doi.org/10.1038/emboj.2011.173; PMID: 21666600
- Ohtani K, Dimmeler S. Epigenetic regulation of cardiovascular differentiation. Cardiovasc Res 2011; 90:404 - 12; http://dx.doi.org/10.1093/cvr/cvr019; PMID: 21372004
- Fish JE, Marsden PA. Endothelial nitric oxide synthase: insight into cell-specific gene regulation in the vascular endothelium. Cell Mol Life Sci 2006; 63:144 - 62; http://dx.doi.org/10.1007/s00018-005-5421-8; PMID: 16416260
- Santhanam L, Christianson DW, Nyhan D, Berkowitz DE. Arginase and vascular aging. J Appl Physiol 2008; 105:1632 - 42; http://dx.doi.org/10.1152/japplphysiol.90627.2008; PMID: 18719233
- Miranda TB, Jones PA. DNA methylation: the nuts and bolts of repression. J Cell Physiol 2007; 213:384 - 90; http://dx.doi.org/10.1002/jcp.21224; PMID: 17708532
- Reik W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature 2007; 447:425 - 32; http://dx.doi.org/10.1038/nature05918; PMID: 17522676
- Casanello P, Krause B, Torres E, Gallardo V, González M, Prieto C, et al. Reduced l-arginine transport and nitric oxide synthesis in human umbilical vein endothelial cells from intrauterine growth restriction pregnancies is not further altered by hypoxia. Placenta 2009; 30:625 - 33; http://dx.doi.org/10.1016/j.placenta.2009.04.010; PMID: 19501907
- Krause BJ, Prieto CP, Muñoz-Urrutia E, San Martín S, Sobrevia L, Casanello P. Role of arginase-2 and eNOS in the differential vascular reactivity and hypoxia-induced endothelial response in umbilical arteries and veins. Placenta 2012; 33:360 - 6; http://dx.doi.org/10.1016/j.placenta.2012.02.006; PMID: 22391327
- Karantzoulis-Fegaras F, Antoniou H, Lai SL, Kulkarni G, D’Abreo C, Wong GK, et al. Characterization of the human endothelial nitric-oxide synthase promoter. J Biol Chem 1999; 274:3076 - 93; http://dx.doi.org/10.1074/jbc.274.5.3076; PMID: 9915847
- Davis ME, Grumbach IM, Fukai T, Cutchins A, Harrison DG. Shear stress regulates endothelial nitric-oxide synthase promoter activity through nuclear factor kappaB binding. J Biol Chem 2004; 279:163 - 8; http://dx.doi.org/10.1074/jbc.M307528200; PMID: 14570928
- Coulet F, Nadaud S, Agrapart M, Soubrier F. Identification of hypoxia-response element in the human endothelial nitric-oxide synthase gene promoter. J Biol Chem 2003; 278:46230 - 40; http://dx.doi.org/10.1074/jbc.M305420200; PMID: 12963737
- Montañez R, Rodríguez-Caso C, Sánchez-Jiménez F, Medina MA. In silico analysis of arginine catabolism as a source of nitric oxide or polyamines in endothelial cells. Amino Acids 2008; 34:223 - 9; http://dx.doi.org/10.1007/s00726-007-0502-7; PMID: 17520329
- Hracsko Z, Hermesz E, Ferencz A, Orvos H, Novak Z, Pal A, et al. Endothelial nitric oxide synthase is up-regulated in the umbilical cord in pregnancies complicated with intrauterine growth retardation. In Vivo 2009; 23:727 - 32; PMID: 19779107
- Myatt L, Eis AL, Brockman DE, Greer IA, Lyall F. Endothelial nitric oxide synthase in placental villous tissue from normal, pre-eclamptic and intrauterine growth restricted pregnancies. Hum Reprod 1997; 12:167 - 72; http://dx.doi.org/10.1093/humrep/12.1.167; PMID: 9043923
- Barut F, Barut A, Gun BD, Kandemir NO, Harma MI, Harma M, et al. Intrauterine growth restriction and placental angiogenesis. Diagn Pathol 2010; 5:24; http://dx.doi.org/10.1186/1746-1596-5-24; PMID: 20412591
- Trampont P, Roudier M, Andrea AM, Nomal N, Mignot TM, Leborgne-Samuel Y, et al. The placental-umbilical unit in sickle cell disease pregnancy: a model for studying in vivo functional adjustments to hypoxia in humans. Hum Pathol 2004; 35:1353 - 9; http://dx.doi.org/10.1016/j.humpath.2004.07.003; PMID: 15668892
- Wang Y, Gu Y, Zhang Y, Lewis DF. Evidence of endothelial dysfunction in preeclampsia: decreased endothelial nitric oxide synthase expression is associated with increased cell permeability in endothelial cells from preeclampsia. Am J Obstet Gynecol 2004; 190:817 - 24; http://dx.doi.org/10.1016/j.ajog.2003.09.049; PMID: 15042020
- Maruyama H, Shinozuka M, Kondoh Y, Akahori Y, Matsuda M, Inoue S, et al. Thrombocytopenia in preterm infants with intrauterine growth restriction. Acta Med Okayama 2008; 62:313 - 7; PMID: 18985091
- le Noble F, Klein C, Tintu A, Pries A, Buschmann I. Neural guidance molecules, tip cells, and mechanical factors in vascular development. Cardiovasc Res 2008; 78:232 - 41; http://dx.doi.org/10.1093/cvr/cvn058; PMID: 18316324
- Ribatti D, Nico B, Crivellato E. Morphological and molecular aspects of physiological vascular morphogenesis. Angiogenesis 2009; 12:101 - 11; http://dx.doi.org/10.1007/s10456-008-9125-1; PMID: 19130273
- Xu XF, Ma XL, Shen Z, Wu XL, Cheng F, Du LZ. Epigenetic regulation of the endothelial nitric oxide synthase gene in persistent pulmonary hypertension of the newborn rat. J Hypertens 2010; 28:2227 - 35; http://dx.doi.org/10.1097/HJH.0b013e32833e08f1; PMID: 20724942
- Godfrey KM, Sheppard A, Gluckman PD, Lillycrop KA, Burdge GC, McLean C, et al. Epigenetic gene promoter methylation at birth is associated with child’s later adiposity. Diabetes 2011; 60:1528 - 34; http://dx.doi.org/10.2337/db10-0979; PMID: 21471513
- Relton CL, Groom A, St Pourcain B, Sayers AE, Swan DC, Embleton ND, et al. DNA methylation patterns in cord blood DNA and body size in childhood. PLoS One 2012; 7:e31821; http://dx.doi.org/10.1371/journal.pone.0031821; PMID: 22431966
- Chan Y, Fish JE, D’Abreo C, Lin S, Robb GB, Teichert AM, et al. The cell-specific expression of endothelial nitric-oxide synthase: a role for DNA methylation. J Biol Chem 2004; 279:35087 - 100; http://dx.doi.org/10.1074/jbc.M405063200; PMID: 15180995
- Bian EB, Huang C, Ma TT, Tao H, Zhang H, Cheng C, et al. DNMT1-mediated PTEN hypermethylation confers hepatic stellate cell activation and liver fibrogenesis in rats. Toxicol Appl Pharmacol 2012; 264:13 - 22; http://dx.doi.org/10.1016/j.taap.2012.06.022; PMID: 22841775
- Lin HY, Kuo YC, Weng YI, Lai IL, Huang TH, Lin SP, et al. Activation of silenced tumor suppressor genes in prostate cancer cells by a novel energy restriction-mimetic agent. Prostate 2012; 72:1767 - 78; http://dx.doi.org/10.1002/pros.22530; PMID: 22539223
- Fish JE, Yan MS, Matouk CC, St Bernard R, Ho JJ, Gavryushova A, et al. Hypoxic repression of endothelial nitric-oxide synthase transcription is coupled with eviction of promoter histones. J Biol Chem 2010; 285:810 - 26; http://dx.doi.org/10.1074/jbc.M109.067868; PMID: 19880524
- Fish JE, Matouk CC, Yeboah E, Bevan SC, Khan M, Patil K, et al. Hypoxia-inducible expression of a natural cis-antisense transcript inhibits endothelial nitric-oxide synthase. J Biol Chem 2007; 282:15652 - 66; http://dx.doi.org/10.1074/jbc.M608318200; PMID: 17403686
- Shirodkar AV, Marsden PA. Epigenetics in cardiovascular disease. Curr Opin Cardiol 2011; 26:209 - 15; http://dx.doi.org/10.1097/HCO.0b013e328345986e; PMID: 21415727
- González RP, Gómez RM, Castro RS, Nien JK, Merino PO, Etchegaray AB, et al. [A national birth weight distribution curve according to gestational age in Chile from 1993 to 2000]. Rev Med Chil 2004; 132:1155 - 65; PMID: 15631202
- Lillycrop KA, Phillips ES, Torrens C, Hanson MA, Jackson AA, Burdge GC. Feeding pregnant rats a protein-restricted diet persistently alters the methylation of specific cytosines in the hepatic PPAR alpha promoter of the offspring. Br J Nutr 2008; 100:278 - 82; http://dx.doi.org/10.1017/S0007114507894438; PMID: 18186951
- Cartharius K, Frech K, Grote K, Klocke B, Haltmeier M, Klingenhoff A, et al. MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics 2005; 21:2933 - 42; http://dx.doi.org/10.1093/bioinformatics/bti473; PMID: 15860560
- Lee JO, Kwun HJ, Jung JK, Choi KH, Min DS, Jang KL. Hepatitis B virus X protein represses E-cadherin expression via activation of DNA methyltransferase 1. Oncogene 2005; 24:6617 - 25; http://dx.doi.org/10.1038/sj.onc.1208827; PMID: 16007161