Abstract
Epigenetic mechanisms precisely regulate sex chromosome inactivation as well as genes that escape the silencing process. In male germ cells, DNA damage response factor RNF8 establishes active epigenetic modifications on the silent sex chromosomes during meiosis, and activates escape genes during a state of sex chromosome-wide silencing in postmeiotic spermatids. During the course of evolution, the gene content of escape genes in postmeiotic spermatids recently diverged on the sex chromosomes. This evolutionary feature mirrors the epigenetic processes of sex chromosomes in germ cells. In this article, we describe how epigenetic processes have helped to shape the evolution of sex chromosome-linked genes. Furthermore, we compare features of escape genes on sex chromosomes in male germ cells to escape genes located on the single X chromosome silenced during X-inactivation in females, clarifying the distinct evolutionary implications between male and female escape genes.
Introduction
The mammalian X and Y chromosomes are believed to have evolved from ordinary autosome pairs,Citation1 and emerged approximately 180 million years ago (MYA) after therian ancestors diverged from the monotreme lineage.Citation2,Citation3 Throughout the course of evolution, the X and Y chromosomes have lost sequence homology due to recurrent gene conversions and Y degeneration. In males, X and Y chromosomes do not have homologous partners. Consequently, a large portion of the X and Y chromosomes does not undergo homologous recombination and remains unsynapsed during meiosis.Citation4 Because of this, the unsynapsed X and Y chromosomes are transcriptionally silenced during male meiosis, and remain silenced into postmeiotic spermatids.Citation5,Citation6 Additionally, in female somatic cells, the genetic disparity of sex chromosomes results in the inactivation of a single X chromosome in order to balance the X chromosome-linked (X-linked) gene dosage between sexes (XX and XY), in a process called X-inactivation.Citation7,Citation8
During this silenced state, a number of sex chromosome-linked (sex-linked) genes are able to escape sex chromosome inactivation and are expressed in both males and females. Due to their tendency to avoid transcriptional silencing, these genes have been named “escape genes”Citation9-Citation12. Although both males and females possess escape genes, the specific “escape genes” identified in female X-inactivation and in male sex chromosome inactivation differ greatly. Emerging evidence has revealed that active genes on the sex chromosomes are precisely regulated in both females and males. In this article, we focus on aspects of escape genes during a state of sex chromosome-wide silencing in postmeiotic spermatids and its relevance to sex chromosome evolution. Notably, profiles of escape genes in postmeiotic spermatids reflect the evolutionary history of mammalian sex chromosomes. By comparing aspects of escape genes found in postmeiotic spermatids to those of inactive female X chromosome, we are able to clarify features of escape genes that are respectively unique to males and females.
Unique Evolutionary Traits of the Sex Chromosomes
Sexual antagonism refers to conflicts that arise between opposing sexes, and is presumed to have impacted the genetic contents of sex chromosomes during the course of evolution. Rice hypothesized that any recessive allele that bestows an advantage to male reproduction is preferentially fixed on the X chromosome, because that allele is readily available via male heterozygosity.Citation13 Due to female homozygosity, female-biased genes have also been proposed to accumulate on X chromosomes. This is because, throughout the course of evolution, the X chromosome has spent twice as much time within females than in males (two Xs in female, but one X in males), and X-linkage of female-biased genes maximizes the opportunity for the female to utilize the allele during the course of evolution.
In the last decade, genome-wide analyses have provided pivotal evidence that supports the accumulation of sexually antagonistic genes on the X chromosome that are expressed in spermatogonia or postmeiotic spermatids.Citation14-Citation16 Comparative analysis of sex chromosomes across species revealed that a class of multi-copy genes located on ampliconic regions of sex chromosomes have recently been acquired, and diverged during mammalian evolution.Citation17-Citation19 Interestingly, this class of genes is predominantly expressed in male germ cells.
The X chromosome is not only rich in sex-biased genes, but also contains many housekeeping genes that are constitutively expressed in both sexes (XX and XY). Conversely, because the Y chromosome is only present in males and is dispensable in females, Y-linked genes are thought to function specifically for male reproduction. Remarkably, the genomic structure and gene content of the Y chromosome are highly divergent across human, chimpanzee and an Old World monkey, rhesus macaque.Citation20 For example, multi-copy genes on the ampliconic region of human Y chromosome display copy number variations between hominoids. Some human Y-linked genes are absent within the chimpanzee genome, suggesting that the genomic contents of the human Y chromosome have been recurrently generated in the past 6 million years.Citation21 Therefore, the male specific function of Y chromosome may be associated with the divergence of its structure and gene content.
Epigenetic Regulation of Sex Chromosomes in Male Germ Cells
In mammals, MSCI is estimated to have emerged nearly 180 MYA, approximately the same time that XY chromosomes emerged after the divergence of the therian ancestor from the monotreme lineage.Citation2 Indeed, MSCI occurs in marsupials,Citation22-Citation24 and its underlying mechanism has been conserved throughout the course of evolution. Accumulating evidence has revealed a significant influence of meiotic sex chromosome inactivation (MSCI) on the evolution of the genome in terms of gene contents and expression.Citation25 During meiosis, while autosomes undergo synapsis and meiotic recombination between homologs, sex chromosomes are largely unsynapsed. Unsynapsed sex chromosomes are transcriptionally silenced by the action of the DNA damage response (DDR) pathway centered on γH2AX and its binding partner MDC1 ().Citation26,Citation27 MSCI is strictly required for male fertility. When MSCI is disrupted, germ cells are arrested at meiotic prophase and are completely eliminated. Recent studies using mice have revealed that hybrid sterility, the phenomenon by which offspring of an inter-species cross become sterile (thereby keeping both species distinct), is associated with disruption of MSCI.Citation28,Citation29 These studies further confirm the strong link between successful MSCI and fertility.
Figure 1. Schematic of epigenetic programming on the sex chromosomes in spermatogenesis of mice. In the pachytene stage, unsynapsed X and Y chromosomes are silenced (MSCI) and form a silent compartment XY body (or sex body). The DDR pathway centered on γH2AX and MDC1 initiates MSCI. Chromosome-wide silencing of the sex chromosomes persists into post-meiotic round spermatids. In the round spermatids, the sex chromosomes occupy a silent compartment, PMSC. Silent epigenetic modifications (CBX1, CBX3, H3K9me3) on the sex chromosomes are maintained from meiosis to round spermatids. The RNF8-dependent active modifications are shown by red bars.
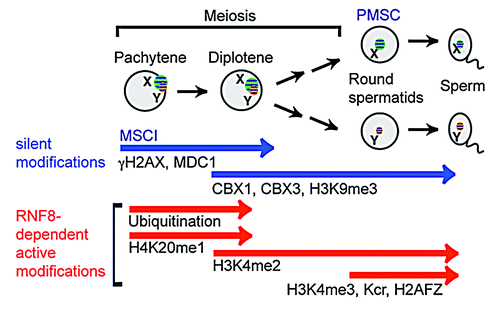
Due to the surveillance of MSCI, gene activation from sex chromosomes is strictly monitored during meiosis. Efficient MSCI results in near complete shut down of sex-linked genes. However, a minor group of genes escape MSCI, this group includes X-linked microRNA genesCitation30 and a non-coding RNA, Tsx.Citation31 Following the initiation of MSCI by the DDR pathway, silent epigenetic modifications (CBX1, CBX3, H3K9me3) are established, and chromosome-wide silencing is established within a silent compartment known as the XY body (or sex body) in spermatocytes. Upon completion of meiosis, silencing is then maintained within postmeiotic sex chromatin (PMSC) found in postmeiotic spermatids.Citation32-Citation34 Despite this transcriptionally repressed state, a group of sex-linked genes escape postmeiotic silencing and become activated on the silent sex chromosomes.Citation10,Citation12,Citation19,Citation32,Citation35
Mechanism Underlying Escape Gene Activation in Postmeiotic Spermatids
A DDR factor, RNF8, has been identified as a regulator of escape gene activation in postmeiotic spermatids.Citation36 RNF8 has been extensively studied in the context of somatic DDR.Citation37-Citation39 RNF8 is an E3 ubiquitin-protein ligase encoding RING finger motif that catalyzes the ubiquitination of histone H2A and H2AX. RNF8-dependent ubiquitination facilitates the recruitment of other DDR factors for subsequent checkpoint activation and DNA repair. In male germ cells, RNF8 mediates ubiquitination of histone H2A on the sex chromosomeCitation40 and subsequently establishes active histone modifications on the sex chromosomes that lead to escape gene activation in postmeiotic spermatids ().
RNF8 is required for the establishment of an active epigenetic modification, H3K4 dimethylation (H3K4me2), on the sex chromosomes during the diplotene phase of meiotic prophase. Because H3K4me2 is the only persistent active mark on the sex chromosome from meiosis to postmeiotic spermatids, H3K4me2 serves as a potential epigenetic memory that leads to escape gene activation in the later stages of postmeiotic spermatids. In postmeiotic spermatids, additional active epigenetic modifications are established in an RNF8-dependent manner on the sex chromosomes. These modifications include H3K4me3, histone lysine crotonylation,Citation41 and histone variant H2AFZ.Citation34 In accordance with the RNF8 dependent establishment of these active epigenetic modifications, escape genes are strongly downregulated in round spermatids of Rnf8 knockout mice. However, a subset of escape genes is not regulated by RNF8, suggesting the possibility of an additional regulatory mechanism of escape gene activation.
In sum, this study represents the first mechanistic insight into the regulation of escape gene activation in postmeiotic spermatids. Importantly, while RNF8 interacts with MDC1 in the same pathway during the somatic DDR, these proteins have distinct functions in male germ cells. RNF8 is associated with gene activation in postmeiotic spermatids, whereas, in contrast, MDC1 is associated with gene silencing in meiosis.Citation27 Therefore, DDR factors have a broad role in epigenetic programming and in controlling gene expression on sex chromosomes in germ cells.
Evolutionary Aspects of Escape Gene Activation in Male Germ Cells
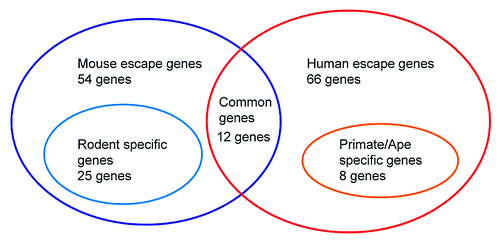
Escape gene activation in postmeiotic spermatids has had a significant impact on the evolution of mammalian sex chromosomes.Citation12 The epigenetic silencing machinery on male sex chromosomes is highly conserved between mice and humans, and the overall gene expression profile of sex-linked genes is consistent between the two species. Most sex-linked genes are stringently repressed at the pachytene stage of meiotic prophase and stay silent in round spermatids, except for escape genes that are mainly associated with male reproductive function. Of note, the gene contents of escape genes are significantly divergent between humans and mice ().Citation12 Of the 66 escape genes found in humans and the 54 found in mice, only 12 escape genes are shared. Of the 54 mice escape genes, 25 are newly evolved genes found only in rodent lineage. Likewise, in humans, 8 out of 66 escape genes are newly evolved genes found only in primate/ape lineages, indicating that gene contents of escape genes are divergent across mammalian species. Together, epigenetic regulation of sex chromosomes is highly conserved, but the content of escape genes is significantly divergent, and those genes tend to be evolutionarily young genes.
Figure 2. Summary of diversity of escape genes on the X chromosome in postmeiotic spermatids. Escape genes in postmeiotic spermatids are significantly diverged between humans and mice. Between the mice and humans (54 escape genes in mice vs 66 escape genes in humans), only 12 escape genes are in common in both groups. Twenty-five out of 54 mice escape genes are newly evolved genes only in rodent lineage, and 8 out of 66 escape genes are newly evolved genes only in primate/ape lineages.
Gene expression changes in spermatids affect the success of fertilization and reproduction. Preferred selection of escape genes in spermatids during the course of evolution might have led to divergent acquisition of escape genes. To test this possibility, the rate of amino acid sequence changes (KA/KS: the ratio of non-synonymous substitutions to the synonymous substitutions) of escape genes between mice and humans were calculated, and compared with that of non-escape genes that are subject to postmeiotic silencing.Citation12 Indeed, the average KA/KS value of escape genes is significantly higher than that of non-escape genes. This result, together with the acquisition of newly evolved escape genes, suggests that escape genes have evolved at a rapid rate in comparison to that of non-escape genes, and that these genes might have been beneficial for reproductive fitness.
What is the evolutionary driving force behind the unique evolution of escape genes? Escape genes in males turned out to be male-biased genes, consistent with Rice’s hypothesis that the fixation of recessive alleles is beneficial to males.Citation13 Therefore, sexual antagonism is a potential driving force that promotes the distribution of these reproduction genes on the sex chromosome in mammals. Divergent acquisition of escape genes may have driven the divergence of reproduction systems among mammalian species. Hence, this feature may have been associated with speciation in mammals. Higher rate of amino acid changes (higher KA/KS value) of escape genes is in line with this notion.Citation12
Genes specifically expressed in round spermatids may affect the success of fertilization and reproduction. As discussed above, the X chromosome should be a preferable location for male reproduction genes; nevertheless, successful sex chromosome inactivation is still necessary to ensure fertility, as demonstrated by the disruption of MSCI in hybrid sterility.Citation28,Citation29
Genomic Strategy against Sex Chromosome Inactivation: Retrotransposition and Multi-Copy Genes
In mammals, an impact of MSCI on genomic evolution is manifested as the retrotransposition of various X-linked genes to autosomes.Citation42,Citation43 Retrotransposition is an efficient mechanism for increasing copy numbers throughout the genome. In the case of genes transposed from the X chromosome to autosomes, X-linked parental genes are generally regarded as housekeeping genes and are ubiquitously expressed. These parental genes on the X chromosomes are silenced during MSCI and silencing is maintained into the postmeiotic stages, while autosomal retrotransposed copies start to express and back up the function of parental genes when MSCI occurs.Citation2,Citation12 Thus, these results suggest that retrotransposition of housekeeping genes is a genomic strategy designed to cope with MSCI.
Another possible strategy involves multi-copy genes that are enriched on sex chromosomes. The structural features of mammalian sex chromosomes are unlike ordinary autosomes. Mammalian sex chromosomes have undergone intra-chromosomal rearrangement, resulting in massive palindromes and ampliconic structures on sex chromosomes. Testis-specific multi-copy genes are enriched in the ampliconic region of sex chromosomes.Citation17-Citation19 Multi-copy genes on mouse X chromosomes are expressed post-meiotically, while most X-linked single copy genes remain repressed during round spermatid stages.Citation19 During the evolutionary past, sexual antagonism might have driven recurrent amplification of multi-copy genes on the sex chromosomes, potentially enabling them to acquire specific post-meiotic expressions against sex chromosome inactivation. For instance, 15 out of 54 escape genes in mice are ascertained as multi-copy genes that were acquired only in rodent lineages. On the other hand, in humans, VCY2 (BPY2), a Y-linked multi-copy escape gene, shows a different copy number when it is compared with our closest relative, chimpanzee, indicating that VCY2 underwent an additional amplification only in human lineage.Citation21 Because of the male-biased expression of this group of genes, hemizygous sex chromosomes possess an advantage to retain these genes on the sex chromosome. Amplified copy number may have increased the chance of being expressed against the barrier of the sex chromosome inactivation. Therefore, the amplification of copy numbers of germ cell genes is a potential genomic strategy designed to cope with postmeiotic silencing.
Intragenomic Conflicts between X- and Y-Linked Multi-Copy Genes
Curiously, sexual antagonism, defined by the conflicts between two sexes, may occur not only between male and female individuals, but could also occur within the genome, between X-bearing sperm (becomes female embryo) and Y-bearing sperm (becomes male embryo). A recent study revealed an example of intragenomic conflicts between X- and Y-linked multi-copy genes in mice.Citation44 Sly, one of the Y-linked multi-copy genes highly expressed in postmeiotic spermatids, has a role in maintaining postmeiotic silencing.Citation45 Its X-linked multi-copy homolog Slx turned out to have an opposite role to that of Sly. Notably, Slx deficiency rescued the phenotype of Sly deficiency, and vice versa, suggesting that conflicts between X-bearing sperm and Y-bearing sperm are regulated by sex-linked multi-copy genes. Furthermore, intragenomic conflicts may contribute to the amplification of the copy number of sex-linked multi-copy genes. It is conceivable that the origin of sexual antagonism could have been the intergenomic conflicts between X-bearing (female) sperm and Y-bearing (male) sperm just after the segregation of X and Y chromosomes during male meiosis.
X Chromosome Inactivation Escape Genes in Females
As mentioned above, a correlation exists between the evolutionary history of sex chromosomes and the profiles of sex-linked escape genes in male germ cells. The regulation of sex chromosomes is distinct between germ cells and somatic cells, and the epigenetic regulation of sex chromosomes in somatic cells also impacts sex chromosome evolution. In female somatic cells, a number of X-linked genes are enabled to escape X inactivation Roughly, 15% of X-linked genes escape X inactivation in humans,Citation46 and 3% of X-linked genes are considered as escape genes in mice.Citation47 A recent study suggests that the number of mouse escape genes can be higher than previously thought, depending on the cell type.Citation48 Thus, in females, the number of escape genes (i.e., degree of escape) is variable in each tissue.
Because females possess a higher dosage of escape genes, potential complications associated with dosage differences of escape genes between females and males may arise. Female escape genes are associated with X aneuploidy syndromes in humans such as Triple X (XXX, 1/1,000 in females), Turner (XO, 1/2,000–1/2,500 in females) and Klinefelter (XXY, 1/500~600 in males) syndromes.Citation49-Citation51 All of the supernumerary X chromosomes are inactivated, leaving only one X chromosome active. Thus, X-linked gene dosage is mostly compensated, but the gene dosage of escape genes is abnormal. A small number of X-linked genes (~5% of X-linked genes) that form large protein complexes are found to be dosage sensitive.Citation52 It would be intriguing to determine whether or not the dosage sensitive X-linked genes that form large protein complexes (identified inCitation52) are functionally responsible for the phenotype of X aneuploidy syndromes.
Curiously, some escape genes from the female inactive X chromosome have homologs on the Y chromosome. Although females have a higher dosage of escape genes due to the inactivation of an entire X chromosome, in males, escape gene dosage may be compensated for by their Y-homologs. However, some studies have reported that there is no direct evidence for the compensation of X-linked genes by Y-linked homologs.Citation53,Citation54 These studies suggest that different gene dosages of female escape genes (those that escape female X inactivation) between female and male somatic tissues may be responsible for phenotypic difference between sexes.
An intriguing difference between escape genes in male germ cells and escape genes in females is their stability during evolution. In female escape genes, the rate of amino acid change remained low throughout the course of evolution (i.e., lower KA/KS value for female escape genes compared with non-escape genes),Citation55 suggesting that strong purifying selection underlies the evolution of female escape genes and that female escape genes are highly conserved. However, as we mention above, male escape genes show higher rates of amino acid change and have significantly diverged for beneficial reproductive fitness. Therefore, female and male escape genes have been derived by distinct evolutionary forces due to the difference in soma vs. germ cell functions.
Concluding Remark
Sex chromosomes have evolved under distinct evolutionary driving forces between somatic cells and germ cells. In an environment of sex chromosome inactivation in germ cells, sexual antagonism could have contributed to the evolution of sex chromosomes in mammals. In males, acquisition of male escape genes from postmeiotic silencing may be associated with higher reproductive fitness, and lead to the divergences of male escape genes in mammals. Because RNF8 is a conserved DDR factor, it would be interesting to explore if the RNF8-dependent mechanism is conserved in mammals other than mice, and if it may give evolutionary pressure for diversification of escape genes. The male sex chromosome inactivation is not only a guardian for genomic integrity of germ cells but potentially contributed to the diversification of genetic contents of sex chromosomes during mammalian evolution.
Acknowledgments
We would like to thank Montserrat Anguera, Eda Yildirim, Yuya Ogawa, Kazuteru Hasegawa for discussion and helpful comments regarding the manuscript; Tyler Broering for editing the manuscript. This work was supported by the Lalor foundation postdoctoral fellowship to H-S.S., the Developmental Fund and Innovation Grant at Cincinnati Children’s Hospital Medical Center to S.H.N., and NIH Grants GM098605 to S.H.N.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Ohno S. Sex chromosomes and sex-linked genes. Berlin, New York [etc.]: Springer-Verlag, 1967.
- Potrzebowski L, Vinckenbosch N, Marques AC, Chalmel F, Jégou B, Kaessmann H. Chromosomal gene movements reflect the recent origin and biology of therian sex chromosomes. PLoS Biol 2008; 6:e80; http://dx.doi.org/10.1371/journal.pbio.0060080; PMID: 18384235
- Veyrunes F, Waters PD, Miethke P, Rens W, McMillan D, Alsop AE, et al. Bird-like sex chromosomes of platypus imply recent origin of mammal sex chromosomes. Genome Res 2008; 18:965 - 73; http://dx.doi.org/10.1101/gr.7101908; PMID: 18463302
- Charlesworth B, Charlesworth D. The degeneration of Y chromosomes. Philos Trans R Soc Lond B Biol Sci 2000; 355:1563 - 72; http://dx.doi.org/10.1098/rstb.2000.0717; PMID: 11127901
- Turner JM. Meiotic sex chromosome inactivation. Development 2007; 134:1823 - 31; http://dx.doi.org/10.1242/dev.000018; PMID: 17329371
- Ichijima Y, Sin HS, Namekawa SH. Sex chromosome inactivation in germ cells: emerging roles of DNA damage response pathways. Cell Mol Life Sci 2012; 69:2559 - 72; http://dx.doi.org/10.1007/s00018-012-0941-5; PMID: 22382926
- Lee JT. Gracefully ageing at 50, X-chromosome inactivation becomes a paradigm for RNA and chromatin control. Nat Rev Mol Cell Biol 2011; 12:815 - 26; http://dx.doi.org/10.1038/nrm3231; PMID: 22108600
- Gendrel AV, Heard E. Fifty years of X-inactivation research. Development 2011; 138:5049 - 55; http://dx.doi.org/10.1242/dev.068320; PMID: 22069183
- Prothero KE, Stahl JM, Carrel L. Dosage compensation and gene expression on the mammalian X chromosome: one plus one does not always equal two. Chromosome Res 2009; 17:637 - 48; http://dx.doi.org/10.1007/s10577-009-9063-9; PMID: 19802704
- Berletch JB, Yang F, Disteche CM. Escape from X inactivation in mice and humans. Genome Biol 2010; 11:213; http://dx.doi.org/10.1186/gb-2010-11-6-213; PMID: 20573260
- Berletch JB, Yang F, Xu J, Carrel L, Disteche CM. Genes that escape from X inactivation. Hum Genet 2011; 130:237 - 45; http://dx.doi.org/10.1007/s00439-011-1011-z; PMID: 21614513
- Sin HS, Ichijima Y, Koh E, Namiki M, Namekawa SH. Human postmeiotic sex chromatin and its impact on sex chromosome evolution. Genome Res 2012; 22:827 - 36; http://dx.doi.org/10.1101/gr.135046.111; PMID: 22375025
- Rice WR. Sex chromosomes and the evolution of sexual dimorphism. Evolution 1984; 38:735 - 42; http://dx.doi.org/10.2307/2408385
- Wang PJ, McCarrey JR, Yang F, Page DC. An abundance of X-linked genes expressed in spermatogonia. Nat Genet 2001; 27:422 - 6; http://dx.doi.org/10.1038/86927; PMID: 11279525
- Khil PP, Smirnova NA, Romanienko PJ, Camerini-Otero RD. The mouse X chromosome is enriched for sex-biased genes not subject to selection by meiotic sex chromosome inactivation. Nat Genet 2004; 36:642 - 6; http://dx.doi.org/10.1038/ng1368; PMID: 15156144
- Zhang YE, Vibranovski MD, Landback P, Marais GA, Long M. Chromosomal redistribution of male-biased genes in mammalian evolution with two bursts of gene gain on the X chromosome. PLoS Biol 2010; 8:8; http://dx.doi.org/10.1371/journal.pbio.1000494; PMID: 20957185
- Skaletsky H, Kuroda-Kawaguchi T, Minx PJ, Cordum HS, Hillier L, Brown LG, et al. The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature 2003; 423:825 - 37; http://dx.doi.org/10.1038/nature01722; PMID: 12815422
- Rozen S, Skaletsky H, Marszalek JD, Minx PJ, Cordum HS, Waterston RH, et al. Abundant gene conversion between arms of palindromes in human and ape Y chromosomes. Nature 2003; 423:873 - 6; http://dx.doi.org/10.1038/nature01723; PMID: 12815433
- Mueller JL, Mahadevaiah SK, Park PJ, Warburton PE, Page DC, Turner JM. The mouse X chromosome is enriched for multicopy testis genes showing postmeiotic expression. Nat Genet 2008; 40:794 - 9; http://dx.doi.org/10.1038/ng.126; PMID: 18454149
- Hughes JF, Skaletsky H, Brown LG, Pyntikova T, Graves T, Fulton RS, et al. Strict evolutionary conservation followed rapid gene loss on human and rhesus Y chromosomes. Nature 2012; 483:82 - 6; http://dx.doi.org/10.1038/nature10843; PMID: 22367542
- Hughes JF, Skaletsky H, Pyntikova T, Graves TA, van Daalen SK, Minx PJ, et al. Chimpanzee and human Y chromosomes are remarkably divergent in structure and gene content. Nature 2010; 463:536 - 9; http://dx.doi.org/10.1038/nature08700; PMID: 20072128
- Namekawa SH, VandeBerg JL, McCarrey JR, Lee JT. Sex chromosome silencing in the marsupial male germ line. Proc Natl Acad Sci U S A 2007; 104:9730 - 5; http://dx.doi.org/10.1073/pnas.0700323104; PMID: 17535928
- Hornecker JL, Samollow PB, Robinson ES, Vandeberg JL, McCarrey JR. Meiotic sex chromosome inactivation in the marsupial Monodelphis domestica. Genesis 2007; 45:696 - 708; http://dx.doi.org/10.1002/dvg.20345; PMID: 17987663
- Franco MJ, Sciurano RB, Solari AJ. Protein immunolocalization supports the presence of identical mechanisms of XY body formation in eutherians and marsupials. Chromosome Res 2007; 15:815 - 24; PMID: 17846907
- Ellegren H. Sex-chromosome evolution: recent progress and the influence of male and female heterogamety. Nat Rev Genet 2011; 12:157 - 66; http://dx.doi.org/10.1038/nrg2948; PMID: 21301475
- Fernandez-Capetillo O, Mahadevaiah SK, Celeste A, Romanienko PJ, Camerini-Otero RD, Bonner WM, et al. H2AX is required for chromatin remodeling and inactivation of sex chromosomes in male mouse meiosis. Dev Cell 2003; 4:497 - 508; http://dx.doi.org/10.1016/S1534-5807(03)00093-5; PMID: 12689589
- Ichijima Y, Ichijima M, Lou Z, Nussenzweig A, Camerini-Otero RD, Chen J, et al. MDC1 directs chromosome-wide silencing of the sex chromosomes in male germ cells. Genes Dev 2011; 25:959 - 71; http://dx.doi.org/10.1101/gad.2030811; PMID: 21536735
- Bhattacharyya T, Gregorova S, Mihola O, Anger M, Sebestova J, Denny P, et al. Mechanistic basis of infertility of mouse intersubspecific hybrids. Proc Natl Acad Sci U S A 2013; 110:E468 - 77; http://dx.doi.org/10.1073/pnas.1219126110; PMID: 23329330
- Campbell P, Good JM, Nachman MW. Meiotic sex chromosome inactivation is disrupted in sterile hybrid male house mice. Genetics 2013; 193:819 - 28; http://dx.doi.org/10.1534/genetics.112.148635; PMID: 23307891
- Song R, Ro S, Michaels JD, Park C, McCarrey JR, Yan W. Many X-linked microRNAs escape meiotic sex chromosome inactivation. Nat Genet 2009; 41:488 - 93; http://dx.doi.org/10.1038/ng.338; PMID: 19305411
- Anguera MC, Ma W, Clift D, Namekawa S, Kelleher RJ 3rd, Lee JT. Tsx produces a long noncoding RNA and has general functions in the germline, stem cells, and brain. PLoS Genet 2011; 7:e1002248; http://dx.doi.org/10.1371/journal.pgen.1002248; PMID: 21912526
- Namekawa SH, Park PJ, Zhang LF, Shima JE, McCarrey JR, Griswold MD, et al. Postmeiotic sex chromatin in the male germline of mice. Curr Biol 2006; 16:660 - 7; http://dx.doi.org/10.1016/j.cub.2006.01.066; PMID: 16581510
- Turner JM, Mahadevaiah SK, Ellis PJ, Mitchell MJ, Burgoyne PS. Pachytene asynapsis drives meiotic sex chromosome inactivation and leads to substantial postmeiotic repression in spermatids. Dev Cell 2006; 10:521 - 9; http://dx.doi.org/10.1016/j.devcel.2006.02.009; PMID: 16580996
- Greaves IK, Rangasamy D, Devoy M, Marshall Graves JA, Tremethick DJ. The X and Y chromosomes assemble into H2A.Z-containing [corrected] facultative heterochromatin [corrected] following meiosis. Mol Cell Biol 2006; 26:5394 - 405; http://dx.doi.org/10.1128/MCB.00519-06; PMID: 16809775
- Wang PJ, Page DC, McCarrey JR. Differential expression of sex-linked and autosomal germ-cell-specific genes during spermatogenesis in the mouse. Hum Mol Genet 2005; 14:2911 - 8; http://dx.doi.org/10.1093/hmg/ddi322; PMID: 16118233
- Sin HS, Barski A, Zhang F, Kartashov AV, Nussenzweig A, Chen J, et al. RNF8 regulates active epigenetic modifications and escape gene activation from inactive sex chromosomes in post-meiotic spermatids. Genes Dev 2012; 26:2737 - 48; http://dx.doi.org/10.1101/gad.202713.112; PMID: 23249736
- Huen MS, Grant R, Manke I, Minn K, Yu X, Yaffe MB, et al. RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell 2007; 131:901 - 14; http://dx.doi.org/10.1016/j.cell.2007.09.041; PMID: 18001825
- Kolas NK, Chapman JR, Nakada S, Ylanko J, Chahwan R, Sweeney FD, et al. Orchestration of the DNA-damage response by the RNF8 ubiquitin ligase. Science 2007; 318:1637 - 40; http://dx.doi.org/10.1126/science.1150034; PMID: 18006705
- Mailand N, Bekker-Jensen S, Faustrup H, Melander F, Bartek J, Lukas C, et al. RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell 2007; 131:887 - 900; http://dx.doi.org/10.1016/j.cell.2007.09.040; PMID: 18001824
- Lu LY, Wu J, Ye L, Gavrilina GB, Saunders TL, Yu X. RNF8-dependent histone modifications regulate nucleosome removal during spermatogenesis. Dev Cell 2010; 18:371 - 84; http://dx.doi.org/10.1016/j.devcel.2010.01.010; PMID: 20153262
- Tan M, Luo H, Lee S, Jin F, Yang JS, Montellier E, et al. Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell 2011; 146:1016 - 28; http://dx.doi.org/10.1016/j.cell.2011.08.008; PMID: 21925322
- Emerson JJ, Kaessmann H, Betrán E, Long M. Extensive gene traffic on the mammalian X chromosome. Science 2004; 303:537 - 40; http://dx.doi.org/10.1126/science.1090042; PMID: 14739461
- Wang PJ. X chromosomes, retrogenes and their role in male reproduction. Trends Endocrinol Metab 2004; 15:79 - 83; http://dx.doi.org/10.1016/j.tem.2004.01.007; PMID: 15036254
- Cocquet J, Ellis PJ, Mahadevaiah SK, Affara NA, Vaiman D, Burgoyne PS. A genetic basis for a postmeiotic X versus Y chromosome intragenomic conflict in the mouse. PLoS Genet 2012; 8:e1002900; http://dx.doi.org/10.1371/journal.pgen.1002900; PMID: 23028340
- Cocquet J, Ellis PJ, Yamauchi Y, Mahadevaiah SK, Affara NA, Ward MA, et al. The multicopy gene Sly represses the sex chromosomes in the male mouse germline after meiosis. PLoS Biol 2009; 7:e1000244; http://dx.doi.org/10.1371/journal.pbio.1000244; PMID: 19918361
- Carrel L, Willard HF. X-inactivation profile reveals extensive variability in X-linked gene expression in females. Nature 2005; 434:400 - 4; http://dx.doi.org/10.1038/nature03479; PMID: 15772666
- Yang F, Babak T, Shendure J, Disteche CM. Global survey of escape from X inactivation by RNA-sequencing in mouse. Genome Res 2010; 20:614 - 22; http://dx.doi.org/10.1101/gr.103200.109; PMID: 20363980
- Calabrese JM, Sun W, Song L, Mugford JW, Williams L, Yee D, et al. Site-specific silencing of regulatory elements as a mechanism of X inactivation. Cell 2012; 151:951 - 63; http://dx.doi.org/10.1016/j.cell.2012.10.037; PMID: 23178118
- Lopes AM, Burgoyne PS, Ojarikre A, Bauer J, Sargent CA, Amorim A, et al. Transcriptional changes in response to X chromosome dosage in the mouse: implications for X inactivation and the molecular basis of Turner Syndrome. BMC Genomics 2010; 11:82; http://dx.doi.org/10.1186/1471-2164-11-82; PMID: 20122165
- Tartaglia NR, Howell S, Sutherland A, Wilson R, Wilson L. A review of trisomy X (47,XXX). Orphanet J Rare Dis 2010; 5:8; http://dx.doi.org/10.1186/1750-1172-5-8; PMID: 20459843
- Tartaglia N, Cordeiro L, Howell S, Wilson R, Janusz J. The spectrum of the behavioral phenotype in boys and adolescents 47,XXY (Klinefelter syndrome). Pediatr Endocrinol Rev 2010; 8:Suppl 1 151 - 9; PMID: 21217607
- Pessia E, Makino T, Bailly-Bechet M, McLysaght A, Marais GA. Mammalian X chromosome inactivation evolved as a dosage-compensation mechanism for dosage-sensitive genes on the X chromosome. Proc Natl Acad Sci U S A 2012; 109:5346 - 51; http://dx.doi.org/10.1073/pnas.1116763109; PMID: 22392987
- Koopman P, Gubbay J, Collignon J, Lovell-Badge R. Zfy gene expression patterns are not compatible with a primary role in mouse sex determination. Nature 1989; 342:940 - 2; http://dx.doi.org/10.1038/342940a0; PMID: 2480529
- Ditton HJ, Zimmer J, Kamp C, Rajpert-De Meyts E, Vogt PH. The AZFa gene DBY (DDX3Y) is widely transcribed but the protein is limited to the male germ cells by translation control. Hum Mol Genet 2004; 13:2333 - 41; http://dx.doi.org/10.1093/hmg/ddh240; PMID: 15294876
- Park C, Carrel L, Makova KD. Strong purifying selection at genes escaping X chromosome inactivation. Mol Biol Evol 2010; 27:2446 - 50; http://dx.doi.org/10.1093/molbev/msq143; PMID: 20534706