Abstract
The metanephric mesenchyme (MM) gives rise to nephrons, the filtering units of the mature kidney. The MM is composed of uninduced (Six2high/Lhx1low) and induced (Wnt-stimulated, Six2low/Lhx1high) cells. The global epigenetic state of MM cells is unknown, partly due to technical difficulty in isolating sufficient numbers of homogenous cell populations. We therefore took advantage of two mouse clonal cell lines representing the uninduced (mK3) and induced (mK4) metanephric mesenchyme (based on gene expression profiles and ability to induce branching of ureteric bud). ChIP-Seq revealed that whereas H3K4me3 active region “peaks” are enriched in metabolic genes, H3K27me3 peaks decorate mesenchyme and epithelial cell fate commitment genes. In uninduced mK3 cells, promoters of “stemness” genes (e.g., Six2, Osr1) are enriched with H3K4me3 peaks; these are lost in induced mK4 cells. ChIP-qPCR confirmed this finding and further demonstrated that G9a/H3K9me2 occupy the promoter region of Six2 in induced cells, consistent with the inactive state of transcription. Conversely, genes that mark the induced epithelialized state (e.g., Lhx1, Pax8), transition from a non-permissive to an active chromatin signature in mK3 vs. mK4 cells, respectively. Importantly, stimulation of Wnt signaling in uninduced mK3 cells provokes an active chromatin state (high H3K4me3, low H3K27me3), recruitment of β-catenin, and loss of pre-bound histone methyltransferase Ezh2 in silent induced genes followed by activation of transcription. We conclude that the chromatin signature of uninduced and induced cells correlates strongly with their gene expression states, suggesting a role of chromatin-based mechanisms in MM cell fate.
Introduction
Histone methylation on lysine or arginine is a key mechanism in regulation of gene expression in development and disease. Among the most important and well-studied nucleosomal modifications are H3K4me3 and H3K27me3. These two histone marks are recognized by distinct sets of transcriptional regulatory complexes and are associated with either activation or repression of gene transcription, respectively.Citation1-Citation3 Interestingly, in the early embryo, bivalent H3K4me3/H3K27me3 domains are frequently found on the same or nearby nucleosomes of silent developmental regulators that are poised for activation and have been implicated in germ cell fate decisions and renewal or differentiation of progenitor cells in developing organs.Citation4-Citation6 Accordingly, there is intense scientific interest in what role chromatin modifications might play in progenitor cell self-renewal and commitment during organogenesis. This information is important in understanding the molecular basis of congenital disease as well as mechanisms of regeneration and repair after injury.
The kidney develops from the intermediate mesoderm via reciprocal interactions between two cell lineages, the metanephric mesenchyme (MM) and the ureteric bud (UB).Citation7-Citation9 A group of MM cells condenses around the UB branch tips to form a nephron progenitor niche, which gives rise to all cell types of the nephron from the glomerulus to the connecting tubule.Citation10 Uninduced nephron progenitors express “stemness” markers such as Six2, Cited1, and Eya1 and are resistant to Wnt-induced differentiation.Citation10-Citation12 In response to inductive Wnt signaling emanating from the adjacent UB, Six2 interacts and cooperates with Lef/Tcf factors and β-catenin to initiate mesenchyme-to-epithelium transition (MET)Citation13,Citation14 and expression of nephrogenic genes such as Wnt4, Lhx1, Pax8, and Fgf8.Citation13,Citation15,Citation16
The present study interrogated the chromatin platform of two clonal mouse cell lines representing the uninduced and induced MM. Whole genome ChIP-Seq and targeted ChIP-qPCR revealed that promoters of silent nephrogenic genes undergo loss of the repressive histone mark, H3K27me3, and/or gain of the active mark, H3K4me3, during differentiation. In contrast, chromatin of identity/renewal genes is depleted of H3K4me3 with reciprocal gain of repressive histone marks. Moreover, stimulation of Wnt/β-catenin signaling in uninduced cells results in a transcriptionally permissive chromatin signature in promoters of nephrogenic genes. Our findings provide new insights into the epigenetic landscape of MM cells.
Results
Genome-wide ChIP-Seq of mK3 and mK4 cell lines
Epigenetic analysis of primary nephron progenitor cells using ChIP technology is a difficult task due to at least two technical reasons: (1) the requirement for a relatively large number of cells; and more importantly, (2) the difficulty in separating the progenitor cells into uninduced and induced populations prior to analysis. Accordingly, we took advantage of two mouse clonal cell lines with gene expression and functional profiles resembling the early uninduced (mK3) and late induced MM (mK4).Citation17 mK3 cells have a spindle-shaped, fibroblast morphology and express genes characteristic of early mesenchyme, including Hoxa11, Hoxd11, collagen I, Twist1, and vimentin, but not epithelial progenitor genes such as Pax2, Wnt4, Pax8 and Lhx1. Moreover, mK3 cells express the nephron stem cell marker Six2, and in organ co-culture experiments they are able to induce growth and branching of the UB. mK4 cells, on the other hand, resemble induced MM undergoing epithelial conversion. These cells are polygonal in shape and express genes characteristic of mesenchyme-to-epithelium transition such as Fgf8, Pax8, Wnt4, Lhx1, HNF1β, E-cadherin and Cadherin-6.
Genome wide ChIP-Seq was performed on mK3 and mK4 cells using specific antibodies to H3K4me3 and H3K27me3, as well as no-antibody control “input.” In all 6 groups, > 15 × 106 unique sequence reads were obtained. To determine areas of significant tag enrichment over background (active regions or peaks), the peak-calling program MACS was used using a p-value of 10−5. The results revealed ~11,000 peaks associated with 10,000 genes (Tables S1 and S2). Genomatix analysis and correlating the number of peaks with distance to the transcription start site (TSS) demonstrates significant enrichment of H3K4me3 and to a lesser extent H3K27me3 around the TSS (). In mK4 cells, the greater the distance from the TSS, the higher the frequency of observed H3K27me3 correlations ().
Figure 1. ChIP-Seq analysis of H3K4me3 and H3K27me3 in uninduced (mK3, yellow) and induced (mK4, blue) metanephric mesenchyme cells.(A and B) Genomatix analysis of the relative distance of H3K4me3 and H3K27me3 active regions (peaks) to the transcription start sites (TSS) of actively transcribed genes in a 20 Kb window. (C and D) Top 5 GO Biological terms determined by GREAT analysis of categorized H3K4me and H3K27me3 active regions (yellow, mK3 unique; blue, mK4; red, shared).
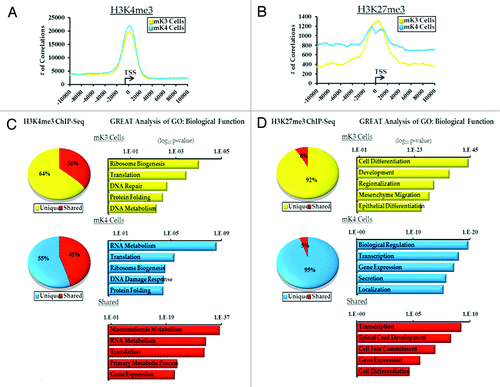
GO Biological Function clustering was performed using the Genomic Regions Enrichment of Annotations (GREAT)Citation18 on all intervals from mK3 and mK4 cells. 55–65% of H3K4me3 intervals are unique to either cell type and 40–50% are shared (). Not surprisingly, since H3K4me3 is found on the promoters of actively transcribed genes, the top categories for all groups contain genes enriched in metabolic function (). In contrast, > 90% of the H3K27me3 intervals are unique to mK3 or mK4 cells. GREAT analysis of these groups found the top categories to be developmental in nature (). Interestingly, mesenchyme migration and epithelial differentiation are among these pathways in mK3 cells in keeping with the biological functions of these cells. The top 5 of each category in GREAT analysis is given in Tables S3–S8, classified by intervals unique to mK3 cells, mK4 cells or common to both.
Distinct chromatin signatures of progenitor and nephrogenic genes
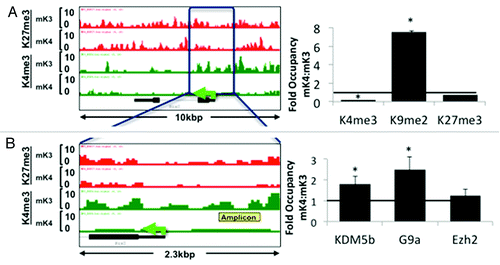
Analysis of ChIP-Seq tracks of more than 100 randomly selected genes as well as of known MM markers differentially expressed in mK3 or mK4 cells was performed using the Integrated Genome Browser (IGB).Citation19 Osr1 and Six2 mark the undifferentiated MM and are not expressed in nascent nephrons; accordingly, they are expressed in mK3 but not mK4 cells. In mK3 cells, Six2 and Osr1 harbor peaks of H3K4me3 around the TSS and broad H3K27me3 intervals. In mK4 cells, these genes are silent and this is associated with loss of H3K4me3 (). ChIP-qPCR was performed on the Six2 5′-regulatory region and confirmed depletion of H3K4me3 as this gene is silenced in mK4 cells (). This was associated with increased occupancy of the H3K4 demethylase Kdm5b and enhanced occupancy of H3K9me2 and its methyltransferase G9a (). Promoter-associated H3K4me3 depletion is a feature shared by most if not all examined progenitor genes that are silenced in mK4 cells.
Figure 2. Chromatin platform of nephron progenitor renewal genes. Snapshots of H3K4me3 (green) and H3K27me3 (red) ChIP-Seq tracks of the progenitor genes Osr1 (A) and Six2 (B) in uninduced (mK3) and induced (mK4) cells. Differentiation is marked by loss of promoter H3K4me3 occupancy.
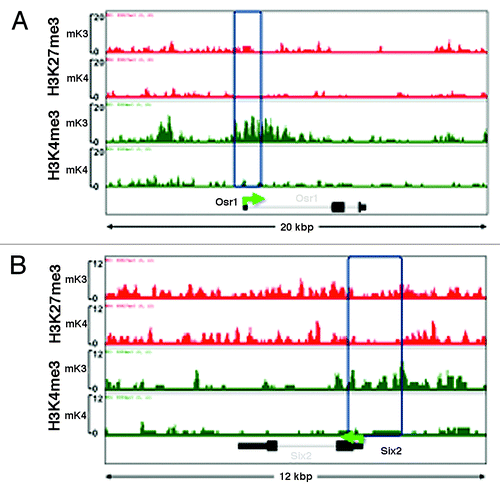
Figure 3.Six2 silencing in induced mK4 cells correlates with acquisition of a repressive chromatin signature. Six2 is expressed in uninduced mK3 but not induced mK4 cells. On the left side, low and high power snapshots of ChIP-Seq H3K4me3 (green) and H3K27me3 (red) tracks are shown. The right panel depicts ChIP-qPCR in the yellow-boxed region. Silencing of Six2 in mK4 cells is associated with loss of H3K4me3, gain of H3K4 demethylase, Kdm5b, and gain of H3K9me2 and methyltrasnferase G9a. Fold occupancy normalized to input and isotype-specific IgG controls. mK3 ChIP value is given a value of 1. n = 3 ChIP experiments per antibody. * p < 0.05 mK4 vs. mK3.
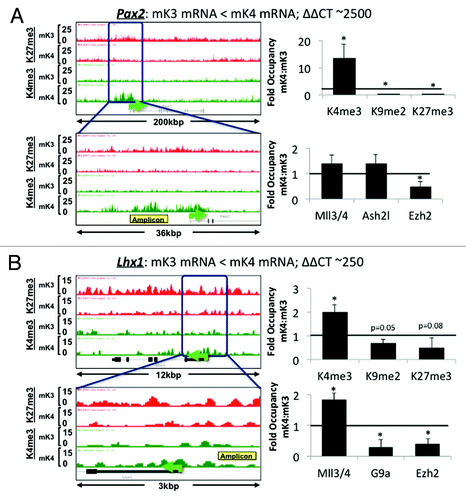
ChIP-Seq analysis revealed that promoters of nephrogenic genes (e.g., Pax8, Jag1, Lef1, Ccnd1) harbor a bivalent chromatin signature in mK3 cells (). In mK4 cells, expression of these genes correlates with gain of H3K4me3, and in some cases, gain of H3K4me3 and loss of H3K27me3 (e.g., Pax2 and Lhx1) (). However, this is not a universal rule; there are examples (e.g., Wnt4, Notch2) where these reciprocal chromatin changes are not so obvious (data not shown), suggesting that other active or repressive marks may be at play. ChIP-qPCR of Pax2 and Lhx1 5′-upstream region in mK3 and mK4 cells confirmed the reciprocal changes occurring in H3K4me3/K27me3 (). This is accompanied by enhanced occupancy of the H3K4 methyltransferase Mll3/4 and reciprocal loss of H3K27 methyltransferase Ezh2. Thus, in general, promoters of nephrogenic genes acquire an active chromatin signature in mK4 cells.
Figure 4. Chromatin platform of nephrogenic genes. Snapshots of H3K4me3 (green) and H3K27me3 (red) ChIP-Seq tracks of genes activated during differentiation. Two major chromatin patterns emerge during differentiation-associated gene activation: (1) loss of repressive H3K27me3 and gain of active H3K4me3 (A,C, and D); or (2) predominant gain of H3K4me3 (B,E, and F).
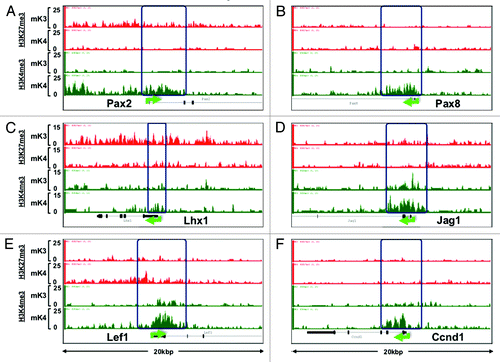
Figure 5. Nephrogenic gene expression correlates with acquisition of active chromatin signatures. On the left side of each panel, low (upper) and high (bottom) power snapshots of ChIP-Seq tracks. The right side of each panel depicts ChIP-qPCR of H3K4me3, H3K9me2 and H3K27me3 and respective modifiers around the yellow-boxed region. In each case, gene activation in mK4 cells is marked by gain of H3K4me3, loss of H3K9me2 and/or H3K27me3 and respective methyltransferases. Fold occupancy normalized to input and isotype-specific IgG controls. ChIP-PCR in mK3 was assigned a value of 1. * p < 0.05 mK4 vs. mK3.
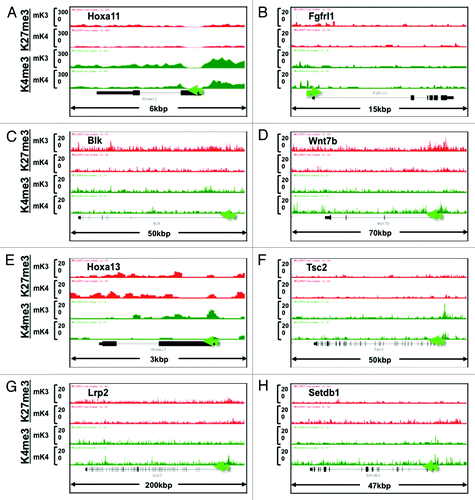
In addition to known kidney developmental genes, we identified a subset of novel progenitor and differentiation genes in mK3 and mK4 cells, respectively, that exhibit a transition in chromatin signature in the silent vs. transcriptionally active state (). Furthermore, we searched the literature for new candidate genes linked to Congenital Anomalies of the Kidney and Urinary Tract (CAKUT)Citation20 and genes associated with glomerular filtration rate.Citation21 shows that the ChIP tracks of these genes conform to one of four patterns in mK3 vs. mK4 cells: high-H3K4me3 in both cell lines (e.g., Hoxa11, Fgfrl1), loss of H3K27me3/gain of H3K4me3 (Blk, Wnt7b), Loss of H3K4me3 (Hoxa13, Tsc2), or gain of H3K4me3 (Lrp2, Setdb1).
Figure 6. Chromatin signature of novel developmental genes expressed in mK3 or mK4 cells. (A–D) Genes expressed in mK3 cells are coated with the active histone mark H3K4me3 around the transcription start site. H3K4me3 peaks are absent when these genes are silent in mK4 cells. (E–H) Genes expressed in mK4 cells are coated with H3K4me3 peaks around the transcription start site. H3K4me3 peaks are absent when these genes are silent in mK3 cells. In the case of Myb (F), there is a net loss of H3K27me3 in mK4 vs. mK3.
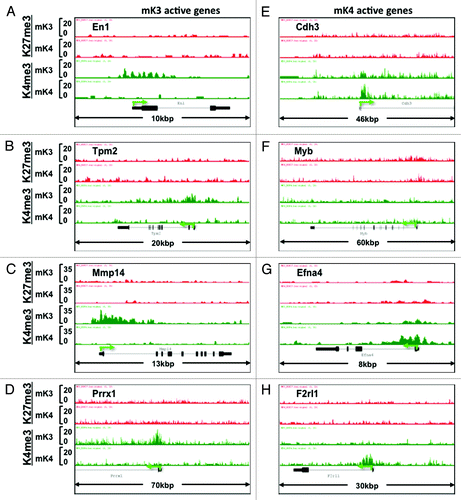
Figure 7. Chromatin signature of candidate developmental genes involved in human CAKUT and in regulation of renal function (see text for details). Transition from mK3 to mK4 cells correlates with gene expression and is associated with several patterns including retention of H3K4me3, loss of H3K27me3/gain of H3K4me3, or gain of H3K4me3 peaks. Loss of H3K4me3 (E and F) correlates with gene repression.
Stimulation of Wnt/β-catenin signaling promotes a permissive chromatin signature in nephrogenic genes
Wnt signaling is the driving force for progenitor cell differentiation in vivo. We examined whether exogenous activation of Wnt signaling in uninduced mK3 cells recapitulates the chromatin changes seen in nephrogenic genes in mK4 cells. mK3 cells were treated with Wnt3a (50 ng/ml) or vehicle (DMSO); RNA was isolated at 6 h and the promoter occupancy was assessed by ChIP-qPCR at 2–4 h. shows that Wnt treatment stimulates expression of nephrogenic genes in mK3 cells and this effect is preceded by enhanced promoter occupancy of H3K4me3 and a reciprocal decrease in H3K27me3. Importantly, these changes in chromatin occupancy correlate with promoter enrichment in β-catenin and a reciprocal decline in Ezh2 occupancy in the same chromatin region (). Thus, we find that promoter recruitment of β-catenin downstream of Wnt signaling to the Tcf/lef binding site is accompanied by loss of pre-bound of Ezh2.
Table 1. Relative enrichment of histone marks, β-catenin and Ezh2 on nephrogenic genes following stimulation of Wnt/β-catenin signaling in undifferentiated MM cells
Discussion
Strong evidence indicates that chromatin states can be predictive of gene expression patterns and cell behavior, such as seen in prostate cancer progressionCitation22 and during germ cell differentiation.Citation23 Genome-wide analysis has revealed that in pluripotent cells, critical genes involved in differentiation, despite remaining silent, have a permissive chromatin structure that makes them sensitive to differentiation-inducing signals.Citation5,Citation24,Citation25 This permissive chromatin environment is characterized by the presence of large H3K27me3 domains harboring peaks of H3K4me3 around the transcriptional start site. Importantly, genes marked with bivalent domains often encode master transcriptional regulators, usually in a lineage-restricted manner, and that are able to orchestrate whole programs of gene expression in differentiated cells.Citation5,Citation26
Our ChIP studies revealed several interesting aspects related to the chromatin states of uninduced and induced MM cells. For example, silencing of the nephron progenitor regulator, Six2, in induced cells correlates not only with loss of promoter H3K4me3 occupancy but also with acquisition of the repressive mark H3K9me2. Moreover, nephrogenic genes undergo dynamic gain and/or loss of activating and repressive chromatin marks, respectively, during differentiation. Interestingly, in developmentally arrested Wilm’s tumor cells, Six2 harbors predominantly the active mark, H3K4me3, whereas Lhx1 is bivalent and silent.Citation27 We also found that activation of Wnt/β-catenin signaling in uninduced cells cells favors a permissive chromatin state on promoters of nephrogenic genes that is characteristic of more differentiated cells, including reciprocal changes in H3K4me3/K27me3, loss of Ezh2, and gain of β-catenin. The bulk of evidence suggests that, depending on the context, H3K27 methylation is a central mechanism to either maintain progenitor cell identity and/or to ensure proper cell renewal and timely differentiation.Citation6,Citation23,Citation28 Our findings agree with a previous report in adipocytes whereby Ezh2 inhibits Wnt signaling.Citation29 Finally, the ChIP-Seq data showed that the majority of candidate genes involved in CAKUT and those linked to GFR exhibit an active chromatin state in mK4 cells (), consistent with a role in mesenchyme-to-epithelium transition. It must be emphasized that our data was derived from two clonal cell lines. Future studies should help confirm these findings in other cell lines and more preferably in primary cells derived from the metanephric mesenchyme. Further understanding of the epigenetic mechanisms of nephron progenitor cell renewal and maintenance is of great clinical importance since low nephron number predisposes to hypertension and chronic kidney disease later in life.Citation30
Materials and Methods
Cell culture
Mouse metanephric mesenchyme cell lines (mK3 and mK4 cells)Citation17 were maintained in media supplemented with 10% fetal bovine serum, penicillin (100 units/ml), and streptomycin (100 µg/ml) at 37°C in a humidified incubator with 5% CO2.
RNA isolation and quantitation of gene expression
Total RNA was isolated using the RNAEasy kit per the manufacturer’s instructions (Qaigen, Valencia, CA). The concentration and purity were determined using NanoDrop 1000/2000 (Thermo Scientific, Wilmington, DE), and diluted to 20 ng/μl. RNA transcripts were measured by quantitative PCR using Tag-Man® primer/probe mixes (Life Technologies Corporation).
ChIP-Seq. ChIP-Seq and input samples were prepared for sequencing using Illumina’s protocol. Briefly, samples were linked to adaptors, library is size-selected (200–250 bp) and PCR-amplified. Sequencing was done on Genome Analyzer 2 (GAII), and the 35-nt sequence reads (“tags”) were mapped to the mouse genome using the ELAND algorithm. The tags were extended unidirectionally in silico at their 3′-ends to a length of 110–200 bp, depending on the average fragment length in the size-selected library. Only tags that map uniquely and have no more than 2 mismatches were used for subsequent analysis. The number of overlapping sequences mapped to a genomic location was determined and visualized in the UCSC genome browser (http://genome.ucsc.edu). H3K4me3 and H3K27me3-enriched chromatin regions (peaks), as defined by MACS, were subjected to GO and GREAT bioinformatics analysis (Tables S1–S8). Real-time quantitative RT-PCR was performed using the TaqMan Gene Expression Assay. Validation of ChIP-Seq data was performed by ChIP-qPCR as described.Citation31-Citation33 mK3 cells were treated with Wnt3a or vehicle (DMSO). ChIP assays for H3K4me3, H3K27me3, β-catenin, and Ezh2 were performed at 2–4 h and quantitative RT-PCR at 6 h post-treatment. Fold change indicates Wnt3a/vehicle ratio. The nephrogenic genes selected have been shown to respond to Wnt signalingCitation34 and are either silent or expressed at very low levels in mK3 cells but are expressed in mK4. *The values are an average of n = 3 experiments for all genes except Etv5 and Sim1 n = 2.
Additional material
Download Zip (1.4 MB)Acknowledgments
The Authors acknowledge Steve Potter (Univ. of Cincinnati) for the mouse metanephric mesenchyme mK3 and mK4 cell lines. ChIP-Seq bioinformatics and statistical analyses were performed in collaboration with GenPathways. This work was supported by NIH grants 1P50 DK096373–01 (Pediatric Center of Excellence in Nephrology) and DK079886, and 1P20 RR017659. We acknowledge the assistance of the Renal and Hypertension Center and The Louisiana Cancer Research Consortium Microscopy and FACS Cores. E.K. is supported by a medical student research grant from the American Society of Nephrology. This work was performed as a part of Ph.D. thesis (N.M.).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental Materials
Supplemental materials may be found here: www.landesbioscience.com/journals/epigenetics/article/25753
References
- van Steensel B, Delrow J, Henikoff S. Chromatin profiling using targeted DNA adenine methyltransferase. Nat Genet 2001; 27:304 - 8; http://dx.doi.org/10.1038/85871; PMID: 11242113
- Handel AE, Ebers GC, Ramagopalan SV. Epigenetics: molecular mechanisms and implications for disease. Trends Mol Med 2010; 16:7 - 16; http://dx.doi.org/10.1016/j.molmed.2009.11.003; PMID: 20022812
- Bonasio R, Tu S, Reinberg D. Molecular signals of epigenetic states. Science 2010; 330:612 - 6; http://dx.doi.org/10.1126/science.1191078; PMID: 21030644
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 2006; 125:315 - 26; http://dx.doi.org/10.1016/j.cell.2006.02.041; PMID: 16630819
- Cui K, Zang C, Roh TY, Schones DE, Childs RW, Peng W, et al. Chromatin signatures in multipotent human hematopoietic stem cells indicate the fate of bivalent genes during differentiation. Cell Stem Cell 2009; 4:80 - 93; http://dx.doi.org/10.1016/j.stem.2008.11.011; PMID: 19128795
- Aldiri I, Vetter ML. PRC2 during vertebrate organogenesis: a complex in transition. Dev Biol 2012; 367:91 - 9; http://dx.doi.org/10.1016/j.ydbio.2012.04.030; PMID: 22565092
- Costantini F, Kopan R. Patterning a complex organ: branching morphogenesis and nephron segmentation in kidney development. Dev Cell 2010; 18:698 - 712; http://dx.doi.org/10.1016/j.devcel.2010.04.008; PMID: 20493806
- Dressler GR. Advances in early kidney specification, development and patterning. Development 2009; 136:3863 - 74; http://dx.doi.org/10.1242/dev.034876; PMID: 19906853
- Little M, Georgas K, Pennisi D, Wilkinson L. Kidney development: two tales of tubulogenesis. Curr Top Dev Biol 2010; 90:193 - 229; http://dx.doi.org/10.1016/S0070-2153(10)90005-7; PMID: 20691850
- Kobayashi A, Valerius MT, Mugford JW, Carroll TJ, Self M, Oliver G, et al. Six2 defines and regulates a multipotent self-renewing nephron progenitor population throughout mammalian kidney development. Cell Stem Cell 2008; 3:169 - 81; http://dx.doi.org/10.1016/j.stem.2008.05.020; PMID: 18682239
- Self M, Lagutin OV, Bowling B, Hendrix J, Cai Y, Dressler GR, et al. Six2 is required for suppression of nephrogenesis and progenitor renewal in the developing kidney. EMBO J 2006; 25:5214 - 28; http://dx.doi.org/10.1038/sj.emboj.7601381; PMID: 17036046
- Brown AC, Muthukrishnan SD, Guay JA, Adams DC, Schafer DA, Fetting JL, et al. Role for compartmentalization in nephron progenitor differentiation. Proc Natl Acad Sci U S A 2013; 110:4640 - 5; http://dx.doi.org/10.1073/pnas.1213971110; PMID: 23487745
- Karner CM, Das A, Ma Z, Self M, Chen C, Lum L, et al. Canonical Wnt9b signaling balances progenitor cell expansion and differentiation during kidney development. Development 2011; 138:1247 - 57; http://dx.doi.org/10.1242/dev.057646; PMID: 21350016
- Park JS, Ma W, O’Brien LL, Chung E, Guo JJ, Cheng JG, et al. Six2 and Wnt regulate self-renewal and commitment of nephron progenitors through shared gene regulatory networks. Dev Cell 2012; 23:637 - 51; http://dx.doi.org/10.1016/j.devcel.2012.07.008; PMID: 22902740
- Carroll TJ, Park JS, Hayashi S, Majumdar A, McMahon AP. Wnt9b plays a central role in the regulation of mesenchymal to epithelial transitions underlying organogenesis of the mammalian urogenital system. Dev Cell 2005; 9:283 - 92; http://dx.doi.org/10.1016/j.devcel.2005.05.016; PMID: 16054034
- Tanigawa S, Wang H, Yang Y, Sharma N, Tarasova N, Ajima R, et al. Wnt4 induces nephronic tubules in metanephric mesenchyme by a non-canonical mechanism. Dev Biol 2011; 352:58 - 69; http://dx.doi.org/10.1016/j.ydbio.2011.01.012; PMID: 21256838
- Valerius MT, Patterson LT, Witte DP, Potter SS. Microarray analysis of novel cell lines representing two stages of metanephric mesenchyme differentiation. Mech Dev 2002; 110:151 - 64; http://dx.doi.org/10.1016/S0925-4773(01)00581-0; PMID: 11744376
- McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, et al. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol 2010; 28:495 - 501; http://dx.doi.org/10.1038/nbt.1630; PMID: 20436461
- Nicol JW, Helt GA, Blanchard SG Jr., Raja A, Loraine AE. The Integrated Genome Browser: free software for distribution and exploration of genome-scale datasets. Bioinformatics 2009; 25:2730 - 1; http://dx.doi.org/10.1093/bioinformatics/btp472; PMID: 19654113
- Sanna-Cherchi S, Kiryluk K, Burgess KE, Bodria M, Sampson MG, Hadley D, et al. Copy-number disorders are a common cause of congenital kidney malformations. Am J Hum Genet 2012; 91:987 - 97; http://dx.doi.org/10.1016/j.ajhg.2012.10.007; PMID: 23159250
- Köttgen A, Pattaro C, Böger CA, Fuchsberger C, Olden M, Glazer NL, et al. New loci associated with kidney function and chronic kidney disease. Nat Genet 2010; 42:376 - 84; http://dx.doi.org/10.1038/ng.568; PMID: 20383146
- Akhtar-Zaidi B, Cowper-Sal-lari R, Corradin O, Saiakhova A, Bartels CF, Balasubramanian D, et al. Epigenomic enhancer profiling defines a signature of colon cancer. Science 2012; 336:736 - 9; http://dx.doi.org/10.1126/science.1217277; PMID: 22499810
- Xu CR, Cole PA, Meyers DJ, Kormish J, Dent S, Zaret KS. Chromatin “prepattern” and histone modifiers in a fate choice for liver and pancreas. Science 2011; 332:963 - 6; http://dx.doi.org/10.1126/science.1202845; PMID: 21596989
- Azuara V, Perry P, Sauer S, Spivakov M, Jørgensen HF, John RM, et al. Chromatin signatures of pluripotent cell lines. Nat Cell Biol 2006; 8:532 - 8; http://dx.doi.org/10.1038/ncb1403; PMID: 16570078
- Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J. A unique chromatin signature uncovers early developmental enhancers in humans. Nature 2010; PMID: 21160473
- Eeckhoute J, Lupien M, Meyer CA, Verzi MP, Shivdasani RA, Liu XS, et al. Cell-type selective chromatin remodeling defines the active subset of FOXA1-bound enhancers. Genome Res 2009; 19:372 - 80; http://dx.doi.org/10.1101/gr.084582.108; PMID: 19129543
- Metsuyanim S, Pode-Shakked N, Schmidt-Ott KM, Keshet G, Rechavi G, Blumental D, et al. Accumulation of malignant renal stem cells is associated with epigenetic changes in normal renal progenitor genes. Stem Cells 2008; 26:1808 - 17; http://dx.doi.org/10.1634/stemcells.2007-0322; PMID: 18467665
- Wyngaarden LA, Delgado-Olguin P, Su IH, Bruneau BG, Hopyan S. Ezh2 regulates anteroposterior axis specification and proximodistal axis elongation in the developing limb. Development 2011; 138:3759 - 67; http://dx.doi.org/10.1242/dev.063180; PMID: 21795281
- Bracken AP, Dietrich N, Pasini D, Hansen KH, Helin K. Genome-wide mapping of Polycomb target genes unravels their roles in cell fate transitions. Genes Dev 2006; 20:1123 - 36; http://dx.doi.org/10.1101/gad.381706; PMID: 16618801
- Mackenzie HS, Lawler EV, Brenner BM. Congenital oligonephropathy: The fetal flaw in essential hypertension?. Kidney Int Suppl 1996; 55:S30 - 4; PMID: 8743507
- Saifudeen Z, Diavolitsis V, Stefkova J, Dipp S, Fan H, El-Dahr SS. Spatiotemporal switch from DeltaNp73 to TAp73 isoforms during nephrogenesis: impact on differentiation gene expression. J Biol Chem 2005; 280:23094 - 102; http://dx.doi.org/10.1074/jbc.M414575200; PMID: 15805112
- Saifudeen Z, Dipp S, Fan H, El-Dahr SS. Combinatorial control of the bradykinin B2 receptor promoter by p53, CREB, KLF-4, and CBP: implications for terminal nephron differentiation. Am J Physiol Renal Physiol 2005; 288:F899 - 909; http://dx.doi.org/10.1152/ajprenal.00370.2004; PMID: 15632413
- Van Bodegom D, Saifudeen Z, Dipp S, Puri S, Magenheimer BS, Calvet JP, et al. The polycystic kidney disease-1 gene is a target for p53-mediated transcriptional repression. J Biol Chem 2006; 281:31234 - 44; http://dx.doi.org/10.1074/jbc.M606510200; PMID: 16931520
- Karner CM, Das A, Ma Z, Self M, Chen C, Lum L, et al. Canonical Wnt9b signaling balances progenitor cell expansion and differentiation during kidney development. Development 2011; 138:1247 - 57; http://dx.doi.org/10.1242/dev.057646; PMID: 21350016