Abstract
In the First German-Catalan Workshop on Epigenetics and Cancer held in Heidelberg, Germany (June 17–19, 2013), cutting-edge laboratories (PEBC, IMPPC, DKFZ, and the Collaborative Research Centre Medical Epigenetics of Freiburg) discussed the latest breakthroughs in the field. The importance of DNA demethylation, non-coding and imprinted genes, metabolic stress, and cell transdifferentiation processes in cancer and non-cancer diseases were addressed in several lectures in a very participative and dynamic atmosphere.
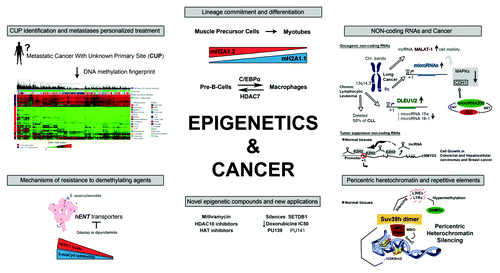
The meeting brought together leading figures in the field of cancer epigenetics to present their research work from the last five years. Experts in different areas of oncology described important advances in colorectal, lung, neuroblastoma, leukemia, and lymphoma cancers. The workshop also provided an interesting forum for pediatrics, and focused on the need to improve the treatment of childhood tumors in order to avoid, as far as possible, brain damage and disruption of activity in areas of high plasticity. From the beginning, the relevance of “omics” and the advances in genome-wide analysis platforms, which allow cancer to be studied in a more comprehensive and inclusive way, was very clear. Modern “omics” offer the possibility of identifying metastases of uncertain origin and establishing epigenetic signatures linked to a specific cluster of patients with a particular prognosis. In this context, invited speakers described novel tumor-associated histone variants and DNA-specific methylation, highlighting their close connection with other processes such as cell-lineage commitment and stemness.
Welcome and Introduction
Angela Risch (German Cancer Research Center, Germany) welcomed participants, acknowledging the close and excellent collaboration between all the research centers and group leaders gathered there. The meeting was organized by epigenetics@dkfz, a cross-program initiative at the German Cancer Research Center (DKFZ), in close collaboration with the Cancer Epigenetics and Biology Program, Bellvitge Biomedical Research Institute, and the Institute of Predictive and Personalized Medicine of Cancer, IMPPC, both in Barcelona, and the CRC 992 Medical Epigenetics at the University of Freiburg. The meeting brought together scientists working in various areas the field of Epigenetics and Cancer, with the aim of developing collaborative efforts and future exchange and training opportunities. Overviews of the epigenetic research activities taking place in their respective centers were provided by Manel Esteller, Director of the Cancer Epigenetics and Biology Program in Barcelona; Manuel Perucho, Director of the Institute of Predictive and Personalized Medicine of Cancer in Barcelona; and Christoph Plass, Head of the Division for Epigenomics and Cancer Risk Factors at the German Cancer Research Center in Heidelberg. The attendees are shown in and the highlights of the meeting are illustrated in .
Novelties in Epigenetic Drugs
This topic was introduced by Olaf Witt (German Cancer Research Center, Germany), who presented data on the selective targeting of histone deacetylases (HDACs) in pediatric oncology. In 2012, his group published a phase I/II trial in children with relapsed solid tumors, leukemias, and lymphomas treated with optimized individualized doses of Vorinostat/SAHA.Citation1 Recent studies by his group have revealed that by targeting histone deacetylase 10, neuroblastoma cells can be sensitized to the chemotherapeutic compounds doxorubicin through a novel discovered mechanism involving dysregulated autophagy by HDAC10.Citation2 These results underscore the need to unmask the molecular functions of single HDAC isozymes to decrease toxicity of unselective inhibition.
Manfred Jung (University of Freiburg, Germany) presented recent efforts made in the field of synthesis and biological testing of novel histone acetyltransferase (HAT) inhibitors.Citation3 For example, pan-HAT inhibitor arose as an effective treatment upon recognition of its ability to suppress acetyltransferase activity in neuroblastoma cells and to block transcription in Schistosomes.
Maria Rius (German Cancer Research Center, Germany) outlined the mechanisms by which patients could become refractory to low doses of 5-azacytidine (5-azaCyd) or 5-aza-2'-deoxycytidine. The uptake of both nucleoside analogs is performed by two types of cell membrane transporters, hCNTCitation4 and hENT. Inhibition of hENT transporter by dilazep or dipyridamole demonstrates that 5-azaCyd-mediated cytotoxicity and DNA demethylation requires its activity. Dr Rius highlighted the importance of correlating the expression and subcellular localization of transport proteins to clinical and molecular response variables from patients undergoing demethylation therapy with 5-azanucleosides, and of evaluating hENT1 expression as a potential biomarker for demethylating agent responses.
Manel Esteller (Cancer Epigenetics and Biology Program, Bellvitge Biomedical Research Institute) concluded the topic, stressing the importance of studying amplifications and deletions of epigenetic genes such as DNMT3B and SETDB1 in cancer, and identifying which epigenetic or non-epigenetic drugs might successfully target them in future clinical applications. Dr Esteller showed how cancer cells harboring DNMT3B amplifications have higher survival rates than those with low gene dosage when they are exposed to demethylating agents.Citation5 Furthermore, the histone methyltransferase (HMT) SETDB1 was shown to be amplified in non-small and small lung cancer cell lines, thereby conferring higher proliferation rates in cell culture and nude mice models. Strikingly, his results have direct clinical implications because an approved antitumor antibiotic called mithramycin was shown to inhibit SETDB1 expression in lung cancer cell lines, diminishing their growth potential.Citation6 Special mention should be made of Dr Esteller’s announcement of the revolutionary test he intends to launch commercially next year. This is based on the capability of DNA methylation to identify cancers of unknown primary origin (CUPs), and will improve the diagnosis and treatment of patients suffering from these unfortunate diseases.
Non-Coding RNAs and Cancer
Angela Risch (German Cancer Research Center, Germany) is investigation epigenetic regulation of GWA-identified lung cancer susceptibility loci. A promoter methylation screen followed by functional studies identified CHRNB4 as a putative oncogene.Citation7 A genome-wide association study also identified copy number variation (CNV) regions associated with risk of early-onset lung cancer, which harbor deregulated genes and microRNAs, which are being further characterized.
Daniel Mertens (German Cancer Research Center, Germany) talked about the complex regulation of the chromosomal band 13q14.3 in hematological diseases and presented some recent observations about the non-coding RNAs localized in this region. In chronic lymphocytic leukemia (CLL), deletion of at least one allele of 13q14 occurs in more than 50% of cases. However, epigenetic alterations such as aberrant DNA methylation also mimic the effects of the deletion, since it disrupts gene expression inside this region. 13q14.3-associated long non-coding genes (lncRNA genes), DLEU1/2, share a CpG island promoter that is methylated in normal cells but is hypomethylated in CLL cells.Citation8 As a result, these lnc-RNAs are upregulated and co-regulate the expression of two neighboring microRNAs (miRNAs), miRNA-15a and miRNA-16–1,Citation9 which are considered to be tumor suppressor genes that are repressed in CLL. Related with this, his studies also revealed defective Drosha-dependent miRNA processing of the aforementioned miRNAs in CLL patients, which results in a higher pri/pre-miRNA ratio. These findings underlined how complex transcription regulation can be in regions that, though they may be considered to be “backstage” of the genome, greatly contribute to the mechanisms driving CLL pathogenesis.
Sven Diederichs (German Cancer Research Center DFKZ and Institute of Pathology, University of Heidelberg, Germany) reported the roles of the lncRNA MALAT-1 in lung adenocarcinoma. This highly conserved and poly(A)-tail lncRNA is deregulated in sarcomas, hepatocellular carcinomas, colon cancer, breast cancer, and other diseases like SARS. Also known as NEAT2, it is highly expressed in lung cancer metastasis and enables us to predict the survival of NSCLC patients. Interestingly, using a novel and efficient technique to obtain knockdown cell lines of MALAT-1 based on the integration of RNA-destabilizing elements through zinc finger nucleases, he observed a substantial reduction in lung cancer metastasis in vitro and in vivo.Citation10,Citation11 He highlighted the use of clinical therapeutic approaches, such as those involving antisense oligonucleotides (ASOs), to prevent lung cancer metastasis.
Dr Esteller gave a general overview of non-coding RNAs in cancer and other diseases,Citation12 drawing particular attention to the roles of those less frequently mentioned, in particularly the PIWI-interacting RNAs (piRNAs), the small nucleolar RNAs (snoRNAs), the transcribed ultraconserved regions (T-UCRs), and the large intergenic non-coding RNAs (lincRNAs). He reported in more detail the epigenetic inactivation of snoRNAs in human cancer, particularly in leukemia,Citation13 and the discovery of a new EZH2-interacting RNA transcribed from an intronic region of the H3K4 methyltransferase gene SMYD3, which has active roles in controlling polycomb guiding and gene repression.Citation14
Thomas Brabletz (Medical Centre, University Freiburg, Germany) described the relevance of the miR-200C family in promoting the EMT and how this can be reverted by a process known as mesenchymal-epithelial transition (MET) in order to establish a distant metastasis. Basically, this family of miRNAs controls which of the two processes prevails over the other through a ZEB1-mediated feedforward loop. Thus, on one hand, ZEB1 expression represses these miRNAs, promoting the EMT in pancreatic and colorectal cancer cells, while on the other, the miR-200C family stimulates epithelial differentiation by ZEB1 downregulation and the limiting of Notch signaling.Citation15 In addition to this bi-directional regulation, miR200-C family expression can also be regulated by methylation of its CpG island promoter.Citation16 In his concluding remarks, he mentioned the appearance of new therapeutic treatments using HDAC inhibitors, which have demonstrated an ability to block efficiently the tumor growth dependent on the miR-200C/ZEB1 cascade in combination with Gemcitabine.
Chromatin Architecture in Cancer: Histones and Variants
Sebastian Bender (German Cancer Research Center, Germany) reported additions to our knowledge of the epigenetics of pediatric high-grade gliomas (pHGGs). Around 30–50% of pHGGs harbor a mutation of H3.3/H3.1 histone (K27M or G34R/V). K27M mutants feature a global reduction in H3K27 trimethylation. This defective protein attaches to the polycomb complex and interferes with the catalytic activity of the EZH2 protein provoking important modifications of gene regulation.
Marcus Buschbeck (Institute of Predictive and Personalized Medicine of Cancer, IMPPC, Barcelona, Spain) talked about chromatin regulation of cell fate transition. Analyses of macroH2A revealed that it is crucial for cell fate transitions including embryonic development of zebrafish,Citation17,Citation18 stem cell differentiationCitation19 and cancer.Citation20 In contrast to the prevailing opinion, they found that macroH2A does not only function as a repressor but also has proactivating functions during embryonic stem cell differentiation.Citation21 How this works on the molecular level is at the present unclear, but the presented but unpublished observation that macroH2A exists in very distinct types of chromatin provides an important clue. An exciting and poorly understood aspect of macroH2A variants is the existence of a metabolite-binding site splice variant of macroH2A1.Citation22 Aiming to understand this function, they presented preliminary data showing a splicing switch during muscle differentiation leading to the upregulation of the metabolite-binding form. These results suggest a possible new function for macroH2A linking metabolic and epigenetic regulation in muscle. Monica Suelves, another member of the IMPPC, also dealt with the muscle differentiation process in her study of its methylation dynamics. She concluded that DNA methylation patterns are highly preserved, although significant changes occur predominantly at CpG-poor/intermediate regions and that there is a loss of methylation during muscle lineage commitment in critical muscle-regulatory genes such as PAX3, MyoD, and Myogenin.
Joan Ausió (University of Victoria, Canada) outlined the role of the histone variant H2A.Z on nucleosome structure and its potential involvement in prostate cancer, focusing on two of its different subtypes, H2A.Z-1 and H2A.Z-2. He emphasized the differential ability of H2A.Z histones to bind to the DNA depending on whether they are in an acetylated or an ubiquitinated state, their homotypic or heterotypic presence in the nucleosome and the potential involvement of the two subtypes. The main focus of his presentation, however, concerned the involvement of H2A.Z in prostate cancer, whereby H2A.Z and its subtypes are involved in the regulation of prostate-specific antigen (PSA) gene transcription, upon androgen stimulation.Citation23
Epigenetic Regulation of Pericentric Heterochromatin (PcH), Mobile Genetic Elements (MGE), and Germ Line Genomes
Karsten Rippe (German Cancer Research Center, Germany) addressed how chromatin controls genome function, particularly through the study of the chromatin surrounding the centromeres. Overall, pericentric heterochromatin (PcH) harbors two main repressive histone marks, H3K9 trimethylation and H4K20 trimethylation. In normal fibroblasts, these marks are established mainly by two HMTs, Suv39h and Suv4–20h, which couple with HP1 and other proteins (e.g., DNMTs) in larger complexes. On the one hand, Suv39h complexes can promote localized propagation of H3K9m3 modifications that direct DNA methylation of satellite repeats at PcH,Citation24 while on the other, Suv4–20h complexes propagate the H4K20m3 histone mark. Interestingly, the losses of these epigenetic imprints increase genome instability, constituting a hallmark for tumor cells.
Aydan Karslioglu (Max-Planck Institute of Immune Biology and Epigenetics, Freiburg, Germany) discussed the regulation of Suv39h-dependent embryonic stem cell (ESC) heterochromatin. This HAT protein promotes deposition of the H3K9m3 repressive histone mark over long interspersed nuclear elements (LINEs) and long-terminal repeats (LTRs). Interestingly, individual copies of these elements produced by defective transposition have few H3K9me3 marks. Her results showed how this process is accomplished by Suv39h, which, in association with HP1 α protein, cooperates to silence LTRs and LINEs.
Manuel Perucho (Institute of Predictive and Personalized Medicine of Cancer, IMPPC, Barcelona, Spain) examined the relation between DNA demethylation of PcH repetitive elements, aging, and the probability of patients developing single or multiple colon cancers. In colorectal cancers, SST1 repeat sequences are not methylated and this accounts for the accumulation of epigenetic errors during cell division in the crypts, especially those affecting chromosomes 4 and 9. This genomic hypomethylation of SST1 is also related to aging and is more common in patients with wild type p53 and genomic damage. The association of DNA demethylation with colon cancer was also confirmed for LINEs against a wild type p53 genetic background. LINE1 demethylation is age-dependent in patients with single cancer (i.e., older age means more demethylation) but is detected at relatively earlier ages in those with multiple cancers. Thus, DNA demethylation has shown its potential as a useful indicator for the clinical management of colon cancer patients, taking into consideration that drastic LINE1 hypomethylation occurs in younger individuals at high risk of having multiple cancers.Citation25
Miguel A. Peinado (Institute of Predictive and Personalized Medicine of Cancer, IMPPC, Barcelona, Spain) recapitulated the DNA methylation remodeling of the Alu repeats, the most abundant family of repeats in the human genome, with over 1 million copies. Interestingly, Alu repeats show higher resistance to hypomethylation than mammalian interspersed repetitive (MIR) elements or LINEs. In fact, hypermethylation of Alu repeats is the norm in normal tissues, but in some cancer types, such as colon adenocarcinomas, Alu repeats are usually unmethylated, contributing to the pathology.
Tanya Vavouri (Institute of Predictive and Personalized Medicine of Cancer, IMPPC, Barcelona, Spain) focused on the chromatin organization of the male germline. Base composition, especially CG- and CpG-content, may influence chromatin architecture in sperm, retaining nucleosomes preferentially around widely expressed and developmental regulatory genes. Her results suggest that nucleosome retention in sperm likely protects these critical regions in the early embryo from epigenetic reprogramming.Citation26 In addition, she briefly reported on the small RNA content of human sperm, which contains a large repertoire of potentially functional regulatory noncoding RNAs, especially RNAs associated with repeats.
Epigenetics of Hematopoietic Transdifferentiation
Esteban Ballestar (Cancer Epigenetics and Biology Program, Bellvitge Biomedical Research Institute) opened the discussion about hematopoietic differentiation by discussing the interplay between transcription factors (TFs) and epigenetic machinery and their impact on the acquisition of targeted epigenetic changes. Two models were discussed: the first one based on C/EBP α-mediated conversion of pre-B cells in macrophages, the second model is the interleukin-stimulated differentiation of monocytes to osteoclasts. In the first model, C/EBP α is able to orchestrate the conversion of the transcription profile without DNA methylation changes.Citation27 However, C/EBP α induces TET2 upregulation and facilitates derepression of myeloid target genesCitation28 without a change in DNA methylation. In the second model, DNA methylation analysis during monocyte to osteoclast differentiation led to the identification of thousands of CpG sites. In this model, dissection of the groups of genes and sequences displaying DNA methylation allows reconstructing the participation of TF and the identification of key interactions between transcription factors and elements of the DNA methylation machinery.
Maribel Parra (Cancer Epigenetics and Biology Program, Bellvitge Biomedical Research Institute) presented recent findings about the key role of HDAC7 in B cell biology, B cell leukemia, and lymphoma. HDAC7 has emerged as a transcriptional repressor of critical genes for macrophage function in B cell precursors (pre-B cells).Citation29 Using an immune reprogramming system she reported that HDAC7 expression is dramatically downregulated during the conversion of pre-B cells into macrophages. Forced expression of HDAC7 interferes with both the acquisition of the macrophage-specific gene signature and the functions of the reprogrammed macrophages. In addition, the model of hematopoietic transdifferentiation serves to identify an epigenetic gene that not only affects pre-B cell identity but also may play a key role in B cell leukemia and lymphoma tumor growth.
Non-Cancer Epigenetics
Dr Esteller summarized the influence of epigenetics on non-cancer pathologies, focusing particularly on neurodegenerative and aging-related diseases. The brain is the most complex of human organs and we still know little about its functionally and failures from an epigenetic and genetic point of view. He reported results showing that each of the brain compartments can be characterized by a specific DNA methylation pattern. Furthermore, although the DNA methylation changes in defective brains vs. normal ones are of low grade compared with what happens in cancer, they are sufficient to induce gene expression changes. Strikingly, he also presented preliminary results that directly connect these DNA methylation modifications with neurodegenerative conditions, such as Alzheimer disease. In addition, great advances are being made in other diseases with a large neurodegenerative component, such as Rett syndrome, which predominantly affects women. In Rett syndrome, loss of Mecp2 transcriptional repressor by mutation deregulates the transcriptional silencing throughout the genome. One of the epigenetic causes of the disease is the upregulation of two lncRNAs, which mediate the downregulation of the gamma-aminobutyric acid receptor subunit Rho 2Citation30 gene, which is associated with susceptibility to epilepsy, one of the side effects of Rett syndrome.
Alejandro Vaquero (Cancer Epigenetics and Biology Program, Bellvitge Biomedical Research Institute) told the audience about the molecular basis of a special family of proteins called sirtuins. These are associated with metabolic and oxidative stress, which are characteristics ultimately related to cancer and aging. SIRT2 was shown to deacetylate H4K16 histone. Its deficiency increases the global levels of H4K16 acetylation, making the chromatin more compact and provoking more errors during mitosis. In addition, H4K16ac coexists with H4K20 monomethylations, whose depositions over the chromatin are stimulated by SIRT2.Citation31 Non-expression of SIRT2 reduces the overall mitotic deposition of H4K20me1 by a mechanism controlled by PR-Set7, thereby impairing genome stability. Curiously, PR-Set7 chromatin localization depends on acetylation by SIRT2. Taken together, the results suggest the direct involvement of SIRT2 in establishing a hitherto unknown mitotic checkpoint.
Dave Monk (Cancer Epigenetics and Biology Program, Bellvitge Biomedical Research Institute) shared with the audience his recent discoveries in the field of genomic imprinting, which is the main contributor to embryonic deficiencies such as the Beckwith–Wiedemann, Silver–Russell, Angelman and Prader–Willi syndromes, among others. Genomic imprinting is the parent-of-origin-specific monoallelic transcriptional silencing observed in placental mammals and is usually driven by DNA methylation. His group recently profiled 33 previously identified differentially methylated regions (DMRs), 8 novel DMRs, and 6 new imprinted domains in patients with imprinting defects using modern platforms such as the Infinium HumanMethylation450 array and the Wide Genome Bisulfite Sequencing process. However, he pointed out that further analyses should be performed to reveal the involvement of these regions in these syndromes. He also argued for the importance of unmasking the mechanisms behind the protection from pre-implantation epigenetic reprogramming. Given that the genome-wide epigenetic states of pre-implantation embryos undergo global reprogramming, he concluded that important mechanisms that are still not well understood must be activated to protect specific areas of the genome from demethylation.
Acknowledgments
The workshop was funded by “epigenetics@dkfz” a cross-program topic at the German Cancer Research Center.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Witt O, Milde T, Deubzer HE, Oehme I, Witt R, Kulozik A, et al. Phase I/II intra-patient dose escalation study of vorinostat in children with relapsed solid tumor, lymphoma or leukemia. Klin Padiatr 2012; 224:398 - 403; http://dx.doi.org/10.1055/s-0032-1323692; PMID: 22915450
- Oehme I, Linke JP, Böck BC, Milde T, Lodrini M, Hartenstein B, et al. Histone deacetylase 10 promotes autophagy-mediated cell survival. Proc Natl Acad Sci U S A 2013; 110:E2592 - 601; http://dx.doi.org/10.1073/pnas.1300113110; PMID: 23801752
- Furdas SD, Shekfeh S, Bissinger EM, Wagner JM, Schlimme S, Valkov V, et al. Synthesis and biological testing of novel pyridoisothiazolones as histone acetyltransferase inhibitors. Bioorg Med Chem 2011; 19:3678 - 89; http://dx.doi.org/10.1016/j.bmc.2011.01.063; PMID: 21353783
- Rius M, Stresemann C, Keller D, Brom M, Schirrmacher E, Keppler D, et al. Human concentrative nucleoside transporter 1-mediated uptake of 5-azacytidine enhances DNA demethylation. Mol Cancer Ther 2009; 8:225 - 31; http://dx.doi.org/10.1158/1535-7163.MCT-08-0743; PMID: 19139132
- Simó-Riudalbas L, Melo SA, Esteller M. DNMT3B gene amplification predicts resistance to DNA demethylating drugs. Genes Chromosomes Cancer 2011; 50:527 - 34; http://dx.doi.org/10.1002/gcc.20877; PMID: 21484930
- Rodriguez-Paredes M, Martinez de Paz A, Simó-Riudalbas L, Sayols S, Moutinho C, Moran S, et al. Gene amplification of the histone methyltransferase SETDB1 contributes to human lung tumorigenesis. Oncogene 2013; In press http://dx.doi.org/10.1038/onc.2013.239; PMID: 23770855
- Scherf DB, Sarkisyan N, Jacobsson H, Claus R, Bermejo JL, Peil B, et al. Epigenetic screen identifies genotype-specific promoter DNA methylation and oncogenic potential of CHRNB4. Oncogene 2013; 32:3329 - 38; http://dx.doi.org/10.1038/onc.2012.344; PMID: 22945651
- Garding A, Bhattacharya N, Claus R, Ruppel M, Tschuch C, Filarsky K, et al. Epigenetic upregulation of lncRNAs at 13q14.3 in leukemia is linked to the In Cis downregulation of a gene cluster that targets NF-kB. PLoS Genet 2013; 9:e1003373; http://dx.doi.org/10.1371/journal.pgen.1003373; PMID: 23593011
- Mertens D, Philippen A, Ruppel M, Allegra D, Bhattacharya N, Tschuch C, et al. Chronic lymphocytic leukemia and 13q14: miRs and more. Leuk Lymphoma 2009; 50:502 - 5; http://dx.doi.org/10.1080/10428190902763509; PMID: 19347735
- Gutschner T, Baas M, Diederichs S. Noncoding RNA gene silencing through genomic integration of RNA destabilizing elements using zinc finger nucleases. Genome Res 2011; 21:1944 - 54; http://dx.doi.org/10.1101/gr.122358.111; PMID: 21844124
- Gutschner T, Hämmerle M, Eissmann M, Hsu J, Kim Y, Hung G, et al. The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res 2013; 73:1180 - 9; http://dx.doi.org/10.1158/0008-5472.CAN-12-2850; PMID: 23243023
- Esteller M. Non-coding RNAs in human disease. Nat Rev Genet 2011; 12:861 - 74; http://dx.doi.org/10.1038/nrg3074; PMID: 22094949
- Ferreira HJ, Heyn H, Moutinho C, Esteller M. CpG island hypermethylation-associated silencing of small nucleolar RNAs in human cancer. RNA Biol 2012; 9:881 - 90; http://dx.doi.org/10.4161/rna.19353; PMID: 22617881
- Guil S, Soler M, Portela A, Carrère J, Fonalleras E, Gómez A, et al. Intronic RNAs mediate EZH2 regulation of epigenetic targets. Nat Struct Mol Biol 2012; 19:664 - 70; http://dx.doi.org/10.1038/nsmb.2315; PMID: 22659877
- Brabletz S, Bajdak K, Meidhof S, Burk U, Niedermann G, Firat E, et al. The ZEB1/miR-200 feedback loop controls Notch signalling in cancer cells. EMBO J 2011; 30:770 - 82; http://dx.doi.org/10.1038/emboj.2010.349; PMID: 21224848
- Davalos V, Moutinho C, Villanueva A, Boque R, Silva P, Carneiro F, et al. Dynamic epigenetic regulation of the microRNA-200 family mediates epithelial and mesenchymal transitions in human tumorigenesis. Oncogene 2012; 31:2062 - 74; http://dx.doi.org/10.1038/onc.2011.383; PMID: 21874049
- Buschbeck M, Uribesalgo I, Wibowo I, Rué P, Martin D, Gutierrez A, et al. The histone variant macroH2A is an epigenetic regulator of key developmental genes. Nat Struct Mol Biol 2009; 16:1074 - 9; http://dx.doi.org/10.1038/nsmb.1665; PMID: 19734898
- Buschbeck M, Di Croce L. Approaching the molecular and physiological function of macroH2A variants. Epigenetics 2010; 5:118 - 23; http://dx.doi.org/10.4161/epi.5.2.11076; PMID: 20160488
- Creppe C, Janich P, Cantariño N, Noguera M, Valero V, Musulén E, et al. MacroH2A1 regulates the balance between self-renewal and differentiation commitment in embryonic and adult stem cells. Mol Cell Biol 2012; 32:1442 - 52; http://dx.doi.org/10.1128/MCB.06323-11; PMID: 22331466
- Cantariño N, Douet J, Buschbeck M. MacroH2A - An epigenetic regulator of cancer. Cancer Lett 2013; 336:247 - 52; http://dx.doi.org/10.1016/j.canlet.2013.03.022; PMID: 23531411
- Creppe C, Posavec M, Douet J, Buschbeck M. MacroH2A in stem cells: a story beyond gene repression. Epigenomics 2012; 4:221 - 7; http://dx.doi.org/10.2217/epi.12.8; PMID: 22449192
- Posavec M, Timinszky G, Buschbeck M. Macro domains as metabolite sensors on chromatin. Cell Mol Life Sci 2013; 70:1509 - 24; http://dx.doi.org/10.1007/s00018-013-1294-4; PMID: 23455074
- Dryhurst D, McMullen B, Fazli L, Rennie PS, Ausió J. Histone H2A.Z prepares the prostate specific antigen (PSA) gene for androgen receptor-mediated transcription and is upregulated in a model of prostate cancer progression. Cancer Lett 2012; 315:38 - 47; http://dx.doi.org/10.1016/j.canlet.2011.10.003; PMID: 22055461
- Lehnertz B, Ueda Y, Derijck AA, Braunschweig U, Perez-Burgos L, Kubicek S, et al. Suv39h-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin. Curr Biol 2003; 13:1192 - 200; http://dx.doi.org/10.1016/S0960-9822(03)00432-9; PMID: 12867029
- Kamiyama H, Suzuki K, Maeda T, Koizumi K, Miyaki Y, Okada S, et al. DNA demethylation in normal colon tissue predicts predisposition to multiple cancers. Oncogene 2012; 31:5029 - 37; http://dx.doi.org/10.1038/onc.2011.652; PMID: 22310288
- Vavouri T, Lehner B. Chromatin organization in sperm may be the major functional consequence of base composition variation in the human genome. PLoS Genet 2011; 7:e1002036; http://dx.doi.org/10.1371/journal.pgen.1002036; PMID: 21490963
- Rodríguez-Ubreva J, Ciudad L, Gómez-Cabrero D, Parra M, Bussmann LH, di Tullio A, et al. Pre-B cell to macrophage transdifferentiation without significant promoter DNA methylation changes. Nucleic Acids Res 2012; 40:1954 - 68; http://dx.doi.org/10.1093/nar/gkr1015; PMID: 22086955
- Kallin EM, Rodríguez-Ubreva J, Christensen J, Cimmino L, Aifantis I, Helin K, et al. Tet2 facilitates the derepression of myeloid target genes during CEBPα-induced transdifferentiation of pre-B cells. Mol Cell 2012; 48:266 - 76; http://dx.doi.org/10.1016/j.molcel.2012.08.007; PMID: 22981865
- Barneda-Zahonero B, Román-González L, Collazo O, Rafati H, Islam AB, Bussmann LH, et al. HDAC7 is a repressor of myeloid genes whose downregulation is required for transdifferentiation of pre-B cells into macrophages. PLoS Genet 2013; 9:e1003503; http://dx.doi.org/10.1371/journal.pgen.1003503; PMID: 23696748
- Petazzi P, Sandoval J, Szczesna K, Jorge OC, Roa L, Sayols S, et al. Dysregulation of the long non-coding RNA transcriptome in a Rett syndrome mouse model. RNA Biol 2013; 10:10; http://dx.doi.org/10.4161/rna.24286; PMID: 23611944
- Serrano L, Martínez-Redondo P, Marazuela-Duque A, Vazquez BN, Dooley SJ, Voigt P, et al. The tumor suppressor SirT2 regulates cell cycle progression and genome stability by modulating the mitotic deposition of H4K20 methylation. Genes Dev 2013; 27:639 - 53; http://dx.doi.org/10.1101/gad.211342.112; PMID: 23468428