Abstract
We have developed a rapid and sensitive quantitative assay for the measurement of individual allelic ratios. This assay minimizes time and labor, the need for special restriction endonuclease enzymes for polymorphic sites, and avoids heteroduplex formation seen with traditional quantitative PCR-based methods. It has improved sensitivity compared to other methods and is capable of distinguishing 1% differences in allelic expression. This assay, termed Pyrosequencing for Imprinted Expression (PIE), involves the use of an intron-crossing PCR primer to generate the first PCR product. We applied the assay to analyze Insulin-like Growth Factor-2 (IGF2) imprinting in both human and mouse prostate tissues.
Introduction
Genomic imprinting is an epigenetic event that generates monoallelic parent-of-origin-specific expression. It is important during development, and disorders in imprinting have been linked to human disease including Prader-Willi, diabetes, and cancer.Citation1,Citation2 The auto-paracrine Insulin-like Growth Factor 2 (IGF2) gene is a frequently studied region that undergoes loss of imprinting (LOI) in these diseases.Citation3,Citation4 Developing sensitive, efficient assays for quantitating imprinting has been an obstacle to progress in the field.
Current methods for evaluating allele-specific expression have a number of limitations. PCR followed by restriction endonuclease digestion is a traditional method,Citation5,Citation6 but the efficiency of restriction endonucleases is incomplete. Mispaired heteroduplex DNA is commonly formed during PCR amplification and cannot be cleaved, resulting in allelic skewing.Citation7 Using polymorphic small tandem repeats (STR) is reliable, but microsatellites are not commonly found in transcribed sequences.Citation8 Allele-specific amplification with multiple primers for specific matches at 3′ nucleotides has been employed, but primers rarely amplify with equal efficiency.Citation9,Citation10 Hot-stop PCR, an assay for linear quantification of allele ratios, is PCR cycle independent, but requires a restriction endonuclease site that recognizes a polymorphism and radioactivity.Citation11 More recently, DNA sequencing combined with Fluorescent primer extension and dideoxynucleotide assay (Flu-PE and SNuPE) have been proposed,Citation12,Citation13 but these are labor intensive and require gel analysis.
Pyrosequencing is a sensitive approach that uses biotin-labeled nucleotides incorporated into DNA to quantitate alleles. It has been used to detect single nucleotide polymorphisms (SNP).Citation14 In this study we evaluated the sensitivity and specificity of pyrosequencing to quantitate allele-specific expression associated with imprinting.
Results and Discussion
Sensitivity and specificity of PIE
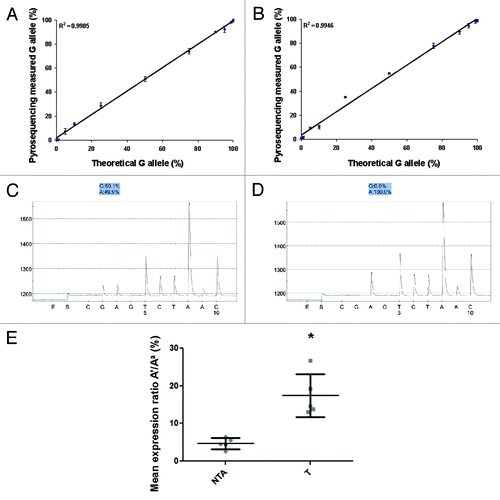
The sensitivity of PIE was evaluated using both human IGF2 and mouse Igf2 SNPs. As described in methods, we varied the number of cycles from 30 to 40 for the nested PCR reaction. The DNA was not well amplified at less than 32 cycles. Employing 35–40 cycle amplification did not alter the ratio between the 2 alleles. PCR products amplified with 35 cycles were used for all experiments described below. As shown in , quantification of the actual allele expression by PIE precisely reflects theoretical expression, as the R-squared value for the trendline is 0.999 and 0.995 for mouse () and human () IGF2, respectively. In both assays, PIE was able to detect 1% differences in allelic ratios. The graphs represent the average value of 3 independent experiments with duplicates within each run. Technical replicates obtained from independent experiments show standard deviations ranging from 0.66–2.52%, indicating a robust assay with negligible inter-PCR and sequencing variation.
Figure 1. (A and B) Detectable range (sensitivity) of Pyrosequencing for Imprinted Expression (PIE). PIE precisely reflected the theoretical ratio of the 2 alleles for both mouse (A) and human (B) IGF2. The trendline showed perfect linearity (R2 > 0.99) for both assays, and 1% changes were able to be detected. The graphs were generated from 3 independent experiments and the data shown as Mean ± SD. (C and D) Accuracy (specificity) of PIE. Pyrosequencing measured equal amounts of both alleles in genomic DNA from heterozygous mice (C), but only demonstrated expression from one allele using mRNA (D) when the intron-crossing primers were used. (E) PIE demonstrates LOI at IGF2 in prostate tissues associated with prostate cancer. Five normal prostates (NTA) with no evidence of inflammation or cancer and 5 prostate tumor tissues (T) were analyzed. Prostate tumor showed significantly LOI of IGF2 compared with NTA tissue. Data are shown as mean ± SD, * t-test p < 0.05.
The same mixed human samples were applied to the Flu-PE assay performed as described.Citation12,Citation15 When the dominant allele appears as the lower band, there is always a ~30% leakage from the recessive (i.e., silenced) allele despite varying primers and conditions (data not shown). The Flu-PE assay for detecting the mouse Igf2 locus exhibits the same problem. PIE does not demonstrate this issue. Furthermore, the completed PIE assay only takes 50% of the time (~8 h) and does not require gel analysis as Flu-PE does.
Next, cDNA and genomic DNA from wild type CI mouse tails were evaluated to check the specificity of PIE. As shown in , using PCR products generated with intron-crossing primers (Method 1), gDNA showed equal percentages (50.1% and 49.9%) from both alleles. The cDNA only showed expression from one allele—the dominant allele A (100%). However, with PCR products generated using primers within the same exon for the 2-step run (Method 2), the cDNA exhibited ~20% expression from the other allele. We believe this to be due to DNA contamination despite several rounds of DNase. The PIE assay with the addition of an intron-crossing primer (Method 1) completely eliminates any non-specific expression from the other allele and is the recommended approach for the assay.
Imprinting status of IGF2 in normal human prostate tissues and prostate cancer tissues
We then analyzed the imprinting status of IGF2 using PIE in 5 normal (NTA) prostate tissues and 5 tumor tissues (T). The NTA showed minimal LOI of IGF2 (2–6.5%) while prostate tumor tissue showed progressive LOI up to 26% (). Data are shown as the recessive allele (A') / dominant allele (Aa) %.
PIE is a new sensitive and efficient assay for the quantification of allelic ratios and is able to detect small differences (in allelic expression). With a strategy of using intron-crossing primers, PIE completely eliminated non-specific expression seen with FLuPE and with heteroduplex formation. The assay is not dependent on PCR cycles and a cycle number greater than 35 did not affect the allelic ratios. This assay does not require a special endonuclease digestion site within the SNP and has much more flexibility to detect any transcribed polymorphism. PIE is a highly reproducible assay with a standard deviation < 2.5% between independent experiments.
One minor limitation is the use of exon-primed intron-crossing (EPIC) primers for the generation of the PCR products, in contrast to using primers within the same exon. The primer can be difficult to generate if a SNP is located in the middle of a very long exon because the primer may not reach the next exon. Intron crossing primers are beneficial, however, because they avoid amplification from the recessive allele due to DNA contamination. This may also happen with other assays when measuring mRNA expression using the primers within the same exon. Researchers have reported that with EPIC-PCR, artifacts such as null alleles are expected to be less frequent than for example, with microsatellites.Citation16,Citation17 We believe that PIE provides an important advance in the detection of imprinting that is rapid, efficient, and reliable and may be applied to a large sample scale.
Materials and Methods
SNP loci
Previously identified SNPs for IGF2 include an A/G polymorphism on exon 5 for human DNA and A/G on exon 6 in the mouse. Previous assays have used ApaI digestionCitation15 and Flu-PE to evaluate allelic expression.Citation12
Tissue samples
Human prostate tissues were obtained from radical prostatectomy patients after approval by the Institutional Review Board at the University of Wisconsin-Madison. Mouse prostate tissues were obtained from the offspring (termed CI) of female C57BL/6 wild type mice crossed with males containing a Mus. Castaneous H19-p57 locus. The CI mouse is thus heterozygous for genotyping (A/G), but only has one paternal allele (A) expressed if the imprint is maintained.
DNA, RNA isolation and cDNA synthesis
Genomic DNA was isolated using the DNeasy Blood and Tissue Kit (Qiagen). RNA was isolated from prostate tissues using RNeasy kit (Qiagen) following the protocol supplied by the manufacturer with the addition of Dnase I to minimize DNA contamination. cDNA was synthesized with the Epitech Reverse Transcription Kit (Qiagen) using 400ng of total RNA. Oligo dTs were used instead of the included RT Primer Mix.
Allelic ratios quantified by pyrosequencing
The PCR products used for pyrosequencing were generated using two methods and the specificity of the two methods was compared. The chosen method was a 2-step PCR approach using exon-primed intron-crossing (EPIC) primers. To do this, a large fragment (1.3kb for human IGF2 and 700bp for mouse Igf2) was amplified using 2μl of cDNA. The primer sequence for human IGF2 was 5'-ATCGTTGAGG AGTGCTGTTT CC (forward) and 5'-GAGCCAGTCT GGGTTGTTGC (reverse) and for mouse Igf2 was 5'-CTCTCAGGCC GTACTTCCGG AC (forward) and 5'-GCGCCGAATT ATTTGATTT (reverse). The amplification program was as follows: initial denaturation of 95 °C for 15 min; 40 cycles of 95 °C for 30 sec, 54 °C for 30 sec and 72 °C for 90 and 45 sec for human and mouse IGF2, respectively. Nested PCR was performed using 0.5 μl of the 1st PCR product with primers flanking SNP regions. These pyrosequencing primers were designed using the PyroMark Assay Design 2.0 software. The pyrosequencing primer sequence for human IGF2 was 5'-AGTCCCTGAA CCAGCAAAGA G (forward) and 5'-TCGGATGGCC AGTTTACC (reverse) and for mouse Igf2 5'-TTCCATCACG TCCCACACTA (forward) and 5'-TGAATATATA ATTTGGGGGG TGTC (reverse). Both reverse primers featured 5' biotin modifications and were HPLC purified, and the final concentration for each primer during PCR reaction was 0.3 µM. Cycling conditions were 15 min of enzyme activation at 95°C followed by 30–40 cycles of 30 sec at 95 °C, 30 sec at 56 °C, and 25 sec at 72 °C.
A second method generated the PCR products directly using the above primers designed for the Pyrosequencing assay within the same exon. Then a nested PCR reaction was performed employing the same primer set.
For both approaches, 5 µL of biotinylated PCR products were captured with Streptavidin sepharose beads, denatured and purified. Then 0.5 µM of pyrosequencing primers (human IGF2: 5'-AGCAAAGAGA AAAGAAGG; mouse Igf2: 5'-AAGGGGATCT CAGCA) were annealed to the purified single-stranded PCR product. Pyrosequencing was performed using PyroMarkTMMD Pyrosequencing System (Qiagen) following the manufacturer’s instructions. The status of each locus was analyzed using PyroMarkTMMD software 1.0 (Qiagen).
For evaluating the imprinting status of normal human prostate tissues and prostate cancer tissues, DNA and RNA from each sample were pyrosequenced simultaneously. Genomic DNA was examined to confirm the heterozygosity (G allele % / A allele % = 1). We then normalized the cDNA according to the ratio obtained from gDNA; LOI is represented as recessive allele (A’) / dominant allele (Aa) %. For each sample, PCR product was generated and pyrosequenced three independent times and run in duplicate format. Statistical analysis was performed by t-test, and p < 0.05 was considered significant.
Detectable range (sensitivity) and accuracy (specificity) of the pyrosequencing assay
To check the minimal detectable range of PIE, a mixing experiment was performed. Two sets of genomic DNA from mouse prostates were employed, each homozygous for different nucleotides at the SNP locus (i.e., G/G and A/A). DNA was mixed in differing ratios ranging from 1% to 100% of a particular nucleotide. Pyrosequencing was subsequently conducted, as described above. This same protocol was used for human DNA.
RNA and genomic DNA from the tails of wild type CI mice were used to calculate the accuracy of pyrosequencing, as these mice are heterozygous at the tested Igf2 locus and retain imprinting in tails.
Acknowledgments
This work was supported by National Institutes of Health, No. 5R01CA097131, (http://grants.nih.gov/grants/)
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Jirtle RL. Genomic imprinting and cancer. Exp Cell Res 1999; 248:18 - 24; http://dx.doi.org/10.1006/excr.1999.4453; PMID: 10094809
- Falls JG, Pulford DJ, Wylie AA, Jirtle RL. Genomic imprinting: implications for human disease. Am J Pathol 1999; 154:635 - 47; http://dx.doi.org/10.1016/S0002-9440(10)65309-6; PMID: 10079240
- Ohlsson R. Loss of IGF2 imprinting: mechanisms and consequences. Novartis Found Symp 2004; 262:108 - 21, discussion 121-4, 265-8; http://dx.doi.org/10.1002/0470869976.ch7; PMID: 15562825
- Ratajczak MZ. Igf2-H19, an imprinted tandem gene, is an important regulator of embryonic development, a guardian of proliferation of adult pluripotent stem cells, a regulator of longevity, and a ‘passkey’ to cancerogenesis. Folia Histochem Cytobiol 2012; 50:171 - 9; http://dx.doi.org/10.5603/FHC.2012.0026; PMID: 22763974
- Wu HK, Squire JA, Catzavelos CG, Weksberg R. Relaxation of imprinting of human insulin-like growth factor II gene, IGF2, in sporadic breast carcinomas. Biochem Biophys Res Commun 1997; 235:123 - 9; http://dx.doi.org/10.1006/bbrc.1997.6744; PMID: 9196048
- Ross JA, Schmidt PT, Perentesis JP, Davies SM. Genomic imprinting of H19 and insulin-like growth factor-2 in pediatric germ cell tumors. Cancer 1999; 85:1389 - 94; http://dx.doi.org/10.1002/(SICI)1097-0142(19990315)85:6<1389::AID-CNCR24>3.0.CO;2-V; PMID: 10189147
- Langhans MT, Palladino MJ. Cleavage of mispaired heteroduplex DNA substrates by numerous restriction enzymes. Curr Issues Mol Biol 2009; 11:1 - 12; PMID: 18541926
- Mansfield ES. Diagnosis of Down syndrome and other aneuploidies using quantitative polymerase chain reaction and small tandem repeat polymorphisms. Hum Mol Genet 1993; 2:43 - 50; http://dx.doi.org/10.1093/hmg/2.1.43; PMID: 8490622
- Pushnova EA, Zhu YS. Quantitative restriction fragment length polymorphism: a procedure for quantitation of diphtheria toxin gene CRM197 allele. Anal Biochem 1998; 260:24 - 9; http://dx.doi.org/10.1006/abio.1998.2682; PMID: 9648648
- Lambertini L, Diplas AI, Lee MJ, Sperling R, Chen J, Wetmur J. A sensitive functional assay reveals frequent loss of genomic imprinting in human placenta. Epigenetics 2008; 3:261 - 9; http://dx.doi.org/10.4161/epi.3.5.6755; PMID: 18769151
- Uejima H, Lee MP, Cui H, Feinberg AP. Hot-stop PCR: a simple and general assay for linear quantitation of allele ratios. Nat Genet 2000; 25:375 - 6; http://dx.doi.org/10.1038/78040; PMID: 10932175
- Fu VX, Dobosy JR, Desotelle JA, Almassi N, Ewald JA, Srinivasan R, et al. Aging and cancer-related loss of insulin-like growth factor 2 imprinting in the mouse and human prostate. Cancer Res 2008; 68:6797 - 802; http://dx.doi.org/10.1158/0008-5472.CAN-08-1714; PMID: 18701505
- Sievers S, Alemazkour K, Zahn S, Perlman EJ, Gillis AJ, Looijenga LH, et al. IGF2/H19 imprinting analysis of human germ cell tumors (GCTs) using the methylation-sensitive single-nucleotide primer extension method reflects the origin of GCTs in different stages of primordial germ cell development. Genes Chromosomes Cancer 2005; 44:256 - 64; http://dx.doi.org/10.1002/gcc.20237; PMID: 16001432
- Royo JL, Galán JJ. Pyrosequencing for SNP genotyping. Methods Mol Biol 2009; 578:123 - 33; http://dx.doi.org/10.1007/978-1-60327-411-1_7; PMID: 19768590
- Fu VX, Schwarze SR, Kenowski ML, Leblanc S, Svaren J, Jarrard DF. A loss of insulin-like growth factor-2 imprinting is modulated by CCCTC-binding factor down-regulation at senescence in human epithelial cells. J Biol Chem 2004; 279:52218 - 26; http://dx.doi.org/10.1074/jbc.M405015200; PMID: 15471867
- Lardeux F, Aliaga C, Tejerina R, Ursic-Bedoya R. Development of Exon-Primed Intron-Crossing (EPIC) PCR primers for the malaria vector Anopheles pseudopunctipennis (Diptera: Culicidae). C R Biol 2012; 335:398 - 405; http://dx.doi.org/10.1016/j.crvi.2012.05.002; PMID: 22721561
- Chenuil A, Hoareau TB, Egea E, Penant G, Rocher C, Aurelle D, et al. An efficient method to find potentially universal population genetic markers, applied to metazoans. BMC Evol Biol 2010; 10:276; http://dx.doi.org/10.1186/1471-2148-10-276; PMID: 20836842