Abstract
Epigenetic events are crucial for early development, but can be influenced by environmental factors, potentially programming the genome for later adverse health outcomes. The insulin-like growth factor 2 (IGF2)/H19 locus is crucial for prenatal growth and the epigenetic state at this locus is environmentally labile. Recent studies have implicated maternal factors, including folate intake and smoking, in the regulation of DNA methylation at this locus, although data are often conflicting in the direction and magnitude of effect. Most studies have focused on single tissues and on one or two differentially-methylated regions (DMRs) regulating IGF2/H19 expression. In this study, we investigated the relationship between multiple shared and non-shared gestational/maternal factors and DNA methylation at four IGF2/H19 DMRs in five newborn cell types from 67 pairs of monozygotic and 49 pairs of dizygotic twins. Data on maternal and non-shared supply line factors were collected during the second and third trimesters of pregnancy and DNA methylation was measured via mass spectrometry using Sequenom MassArray EpiTyper analysis. Our exploratory approach showed that the site of umbilical cord insertion into the placenta in monochorionic twins has the strongest positive association with methylation in all IGF2/H19 DMRs (p < 0.05). Further, evidence for tissue- and locus-specific effects were observed, emphasizing that responsiveness to environmental exposures in utero cannot be generalized across genes and tissues, potentially accounting for the lack of consistency in previous findings. Such complexity in responsiveness to environmental exposures in utero has implications for all epigenetic studies investigating the developmental origins of health and disease.
Introduction
Cellular function in multicellular organisms is mediated by the interplay between underlying genetic profile and epigenetic mechanisms. The most widely studied epigenetic mechanism is DNA methylation that occurs predominately at the cytosine base of the CpG dinucleotide.Citation1,Citation2 The epigenome is in a rapid state of flux during prenatal developmentCitation3 and evidence shows that it is particularly susceptible to environmental influence during this period.Citation4 Following evidence that placenta weight and/or birth weight can convey an elevated risk for cardiovascular and metabolic disease, a number of animal studies and a small number of human studies have since concluded that it is likely that environments that restrict fetal growth may “program” future chronic disease through influence on epigenetic state in utero.Citation5-Citation8
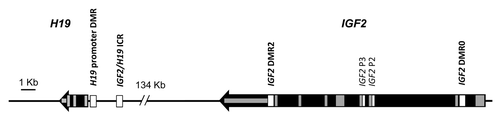
DNA methylation-mediated changes in gene expression play a pivotal role in early embryonic development, through two waves of genome-wide demethylation coupled with selective re-methylation.Citation9-Citation12 Expression of mammalian imprinted genes is controlled by differentially-methylated regions (DMRs), a subset of which act as master imprinting control regions (ICRs) for loci containing two or more imprinted gene.Citation13,Citation14 The two most widely-studied imprinted genes are H19 and IGF2, which are co-regulated within the same locus at human chromosome 11p15 (). Both are important for fetal and placental growth.Citation15 Within this locus, the IGF2/H19 ICR upstream of the H19 gene is a paternally-methylated germline DMR located within a series of binding sites for the CTCF protein necessary for long range chromatin interactions controlling gene activity.Citation16,Citation17 There are at least three other DMRs within the IGF2/H19 locus whose methylation status is somatically acquired during early embryonic development.Citation18,Citation19
Figure 1. Structure of the IGF2/H19 locus with DMRs. The IGF2/H19 locus is shown with direction of transcription, exon locations (gray boxes) and DMRs (white boxes). DMRs assayed for DNA methylation in this study are indicated in bold.
In rodents, gestational environments such as maternal protein restriction,Citation7,Citation20,Citation21 low dietary folate,Citation22-Citation24 low vitamin B12,Citation25,Citation26 high methionineCitation22 and alcohol consumptionCitation27-Citation31 have been shown to influence DNA methylation in offspring, including at the IGF2/H19 locus.Citation28,Citation29 In humans, maternal depression/anxiety,Citation32,Citation33 micronutrient supplements,Citation34 smoking,Citation35,Citation36 famine in early gestation,Citation37,Citation38 periconceptual folic acid intake,Citation39,Citation40 and vitamin B12 intakeCitation41 have also been implicated in the regulation of DNA methylation in offspring, including at the IGF2/H19 locus.Citation34,Citation36,Citation37,Citation39-Citation41 However, previous studies of IGF2/H19 DNA methylation in humans have focused on single tissues, and generally single environmental exposures (with one exception).Citation41
Twins have traditionally been used to calculate the proportion of phenotypic variance explained by variation in genetic, common environmental and unique environmental factors,Citation42 and this approach has begun to pinpoint a role for each in establishing and maintaining levels of DNA methylation.Citation43,Citation44 In our recent studies of newborn twins, we presented evidence that DNA methylation within the IGF2/H19 locusCitation45 and throughout the genomeCitation46 in humans is governed by genetic and non-shared stochastic and/or environmental factors, with latter being the largest component of variation. However, the relative contributions of specific environmental exposures to inter-individual differences in DNA methylation were not examined. Although twins share the same common (maternal) environment, they have individual supply lines, with each twin having its own umbilical cord with associated placental insertion site, placental circulation (with the exception of some monochorionic [MC] pairs) and in > 99% of cases, separate amniotic sacs.Citation47 These factors contribute to a unique intrauterine environment for each twin within a pair. Position of cord insertion into the placenta and uneven placental allocation are significantly associated with birth weight discordance in MC twin pairs.Citation48-Citation50 Velamentous insertion is also associated with increased perinatal morbidity and mortality.Citation51 In velamentous cord insertion, the umbilical cord inserts into the fetal membranes and the umbilical blood vessels then travel, unprotected, within the membranes to the placenta. Here, we provide a detailed analysis of the effects of ten early life factors on DNA methylation throughout the IGF2/H19 locus, in multiple tissues at birth using data collected within the Peri/postnatal Epigenetic Twins Study (PETS).Citation52 Our goal was to explore the association of the shared maternal and non-shared supply line factors with IGF2/H19 methylation at the four loci specified above. We were particularly interested in each measure’s relationship with DNA methylation after taking the effects of the other measures into account, and whether the relationships held generally across the five cell types examined, or only to specific cell types. Shared factors included maternal folate and macronutrient intake, alcohol consumption, while twin-specific factors included placental size and cord insertion site. The tissues examined represent multiple lineages: mesoderm [Human Umbilical Vein Endothelial Cells (HUVECs), Cord Blood Mononuclear Cells (CBMCs) and granulocytes]; ectoderm (buccal epithelium) and extra embryonic ectoderm (placenta).
Results
Subjects and samples
Characteristics of the 67 MZ and 49 DZ twin pairs used in this study are listed in . In total we analyzed DNA methylation in DNA from buccal cells from 95 pairs (55 MZ and 40 DZ), CBMCs from 70 twin pairs (36 MZ and 34 DZ), granulocytes from 51 pairs (27 MZ and 24 DZ), HUVECs from 73 pairs (38 MZ and 35 DZ) and placenta from 67 pairs (34 MZ and 33 DZ).
Table 1. Subject characteristics of twin pairs
Association of nutrient intake and lifestyle measures with DNA methylation
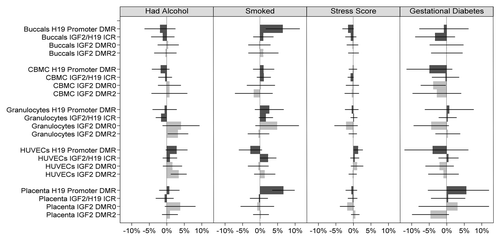
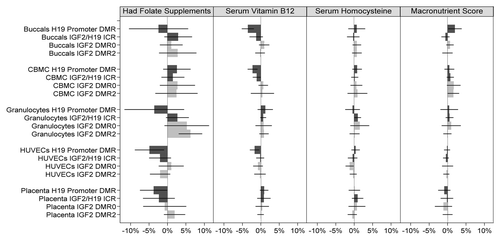
When all factors were taken into account and combining methylation data from all four DMRs (assays) and all five cell types, no significant associations (p < 0.05) were found for nutrient intake and lifestyle measures (All assays combined, ). However, when DMRs were analyzed separately (with cell types still combined), significant associations were observed predominately at the H19 promoter DMR ( and –; Table S2). Newborn offspring of mothers who had supplemental folate in the first 12 weeks of pregnancy exhibited lower mean methylation level compared with newborn offspring of mothers who did not (difference = -1.7%; p = 0.024; 95%CI: -3.2% to -0.2%), and similarly, a 1 standard deviation increase in serum vitamin B12 level at 28 weeks was associated with a -1.0% difference in mean methylation (p = 0.002; 95%CI: -1.6% to -0.4%) at this DMR. On the other hand, a 1 standard deviation increase in macronutrient score was associated with a 0.8% increase in mean methylation (p = 0.049; 95% CI: 0.0% to 1.6%) over all five cell types combined. Newborn offspring of mothers who smoked before they knew they were pregnant had 2.1% higher methylation (p = 0.005; 95% CI: 0.6% to 3.6%) at this locus with all tissues combined. Interestingly, newborn offspring of mothers who smoked also had higher methylation at the IGF2/H19 ICR (difference = 1.2%; p = 0.043; 95% CI: 0.0% to 2.4%) and newborns of mothers who took folate supplementation had higher methylation at the IGF2 DMR2 (difference = 2.9%; p = 0.035; 95% CI: 0.2% to 5.6%; and –; Table S2). Formal testing of differences in associations between loci suggested there may be a difference in the effect of folate supplementation between IGF2 DMR0 and H19 promoter DMR (difference in coefficients = 2.6%; p = 0.034; Table S3) and between IGF2 DMR2 and H19 promoter DMR (difference in coefficients = 4.7%; p = 0.005; Table S3), and a difference in the coefficients of vitamin B12 between IGF2 DMR0 and IGF2/H19 ICR (difference in coefficients = 1.2%; p = 0.023; Table S3). Interestingly, of the observed differences in associations between cell types (Table S4), all except one observed difference was between HUVECs and buccals (folate supplementation; difference in coefficients = -4.5%; p = 0.026; alcohol; 3.0%; p = 0.013; stress score; 1.2%; p = 0.009; Table S4), with the remaining difference being between Granulocytes and Buccals (difference in coefficients = 2.1%; p = 0.004; Table S4).
Table 2. Linear regression of IGF2/H19 methylation jointly on maternal and supply line factors over all five cell types combined
Figure 2. Association of IGF2/H19 methylation and nutrition factors across all cell types and DMRs. Light gray bars represent methylation changes in the IGF2 DMRs, and the dark gray bars represent changes in H19 DMRs. 95% confidence intervals of the coefficient change are represented by a horizontal black line through each bar. The mean methylation differences corresponding to serum vitamin B12, serum homocysteine, and macronutrient are for a 1 standard deviation increase in the respective factor’s z-score. Macronutrient score is obtained as the average of protein, carbohydrate and energy z-scores.
Association of gestational diabetes with DNA methylation
Overall, the presence of gestational diabetes was associated with decreasing DNA methylation when the data were combined (All assays combined, ) and averaged over the four DMRs (assays) and five cell types examined ( and ; Table S2). A difference of -1.3% in mean methylation (p = 0.012; 95%CI: -2.4% to -0.3%) was observed between newborn offspring of mothers with and without gestational diabetes after controlling for the remaining measures (). This negative difference remained consistent across individual loci: -1.5% (H19 promoter DMR), -0.8% (IGF2/H19 ICR), -1.5% (IGF2 DMR0) and -1.6% (IGF2 DMR2). There was no evidence that the association of gestational diabetes with H19/IGF2 methylation differed between loci (Table S3) or between cell types (Table S4).
Association of umbilical cord placement with DNA methylation
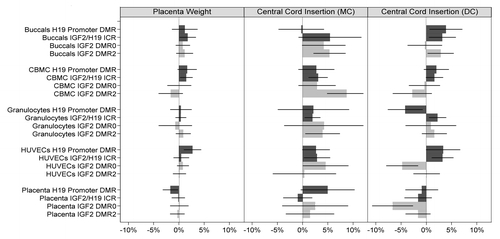
Central location of cord insertion into the placenta for newborns within pairs of MC twins exhibited a positive association with IGF2/H19 methylation when considered over the four loci and five cell types examined. A difference of 3.0% (p < 0.001; 95%CI: 1.6% to 4.4%) in mean methylation between newborns with central cord insertion compared with those with non-central cord insertion was observed after controlling for all other measures examined ( and ; Table S2) in the combined analysis of all assays and cell types (All assays combined, ). The observed difference was consistent at each locus (), namely 3.2% at the H19 promoter DMR (p = 0.046; 95%CI: 0.1% to 6.3%), 2.4% within the IGF2/H19 ICR (p = 0.005; 95%CI: 0.7% to 4.1%), 2.7% at IGF2 DMR0 (p = 0.001; 95%CI: 1.1% to 4.2%) and 4.3% at IGF2 DMR2 (p = 0.002; 95%CI: 1.6% to 7.0%), respectively. Moreover, the observed positive difference remained evident within cell types, with 18/20 tissue-locus pairs exhibiting higher mean methylation in MC newborns with central cord insertion (), the two exceptions being a small negative difference at the H19 promoter DMR in buccals and at the IGF2/H19 ICR in placenta. In contrast, differences in mean methylation between dichorionic (DC) twins with and without central placement of the umbilical cord insertion into the placenta were more varied ( and ; Table S2), with only the IGF2/H19 ICR region exhibiting evidence of a positive difference (of 1.8%; p = 0.001; 95%CI: 0.8% to 2.8%) and the IGF2 DMR0 region exhibiting some evidence of a negative difference (of -1.8%; p = 0.045; 95%CI:-3.6% to 0.0%). The observed difference in association between MC and DC twins may be explainable by a difference in the coefficients of central cord insertion between placenta and non-placenta cell types, with a difference of -4.6% in coefficients between placenta and buccal tissue types being observed (p = 0.003; Table S4).
Figure 4. Association of IGF2/H19 methylation and supply line factors across all cell types and DMRs. Legend as for . Mean methylation differences are regression coefficients obtained from multivariable regression analysis, whereby regression coefficients are converted to percentage difference by multiplying each regression coefficient with average standard deviation. The mean methylation corresponding to placenta weight is for a 1 standard deviation increase in placenta weight z-score.
Association of placenta weight with DNA methylation
Similarly to cord insertion in MC twins, placenta weight exhibited evidence of a positive association with IGF2/H19 methylation when averaged over all four loci and five cell types. An increase of 0.5% in mean methylation per 1 standard deviation increase in placenta weight was observed (All assays combined, ) (p = 0.044; 95%CI: 0.0% to 1.1%), with the positive difference remaining consistent across loci (): 0.5% in the H19 promoter DMR, 0.9% within the IGF2/H19 ICR, 0.3% at IGF2 DMR0 and 0.3% at IGF2 DMR2, respectively ( and ; Table S2). A striking difference between non-placenta and placenta tissues was observed, with 14/16 locus-tissue pairs in non-placenta tissues exhibiting a positive association of placenta weight with DNA methylation (), compared with a negative association of placenta weight with DNA methylation being observed at the H19 promoter region in placenta tissue, and essentially no association being observed at each of the other three loci (; Table S2). Formal testing of differences in coefficients of placenta weight on H19/IGF2 methylation between cell types showed a -1.7% difference (p = 0.032; Table S4) in placenta weight coefficients between placenta and buccals tissue.
Expected antagonistic relationship of IGF2 and H19
IGF2 and H19 work antagonistically to promote and inhibit fetal/placental growth respectively.Citation15,Citation53 As DNA methylation at IGF2/H19 DMRs correlates positively with IGF2 expression, negatively with H19 expression, and is positively correlated with growth, we determined whether the associations observed between maternal and supply line factors (assumed to be also positively associated with growth) and IGF2/H19 DNA methylation were in agreement with these relationships. Eleven of 12 relationships between supply line factors and IGF2/H19 DNA methylation with all cell types combined () had positive regression coefficients as expected; the single exception being central cord insertion in DC twins and DNA methylation at IGF2 DMR0. These positive relationships also held true for 14 out of 18 significant relationships at the DMR and tissue level (, 95%CIs not crossing zero).
Discussion
We have undertaken a comprehensive multivariable study of the association of a range of maternal environmental factors with DNA methylation within IGF2/H19. This is the first study of its kind, encompassing the combined relationship of maternal micronutrient (folate, vitamin B12 and homocysteine), macronutrient, nutrient supply line factors (umbilical cord insertion point and placenta weight), lifestyle factors (alcohol consumption, smoking and perceived stress) and gestational diabetes with DNA methylation levels at multiple loci and across multiple tissues in newborn twins.
The importance of unique environment on the neonatal epigenome
The most striking finding of our study is the observed consistent positive association of central cord insertion of the umbilical cord into the placenta with IGF2/H19 DNA methylation levels in MC twins. This finding suggests the existence of a robust positive association between DNA methylation in the IGF2/H19 region and the quality of the insertion point of the umbilical cord into the placenta for newborns from a pair of MC twins. We also observed a positive association of placenta weight with IGF2/H19 DNA methylation in other tissues. Together, these findings suggest that DNA methylation in this region is susceptible to the quality of the nutrient supply line, but that this is a complex relationship subject to many different influences. In particular, these data suggest that the unique environment experienced individually by each twin of a twin-pair, that includes the differential position of cord insertion and associated placental vascularity, can influence DNA methylation in utero. The reason why an association with cord insertion was observed for only MC twins could reflect the competition for resources that can occur within twin pairs shat share the same placenta.Citation48 As DNA methylation within the IGF2/H19 locus can influence fetal growth through its effects on gene expression,Citation15-Citation17 we suggest that the impeded blood flow that results from a smaller placenta sizeCitation54 or a non-central cord insertion pointCitation55 influences fetal growth at least in part due to an increase in DNA methylation within IGF2/H19. Indeed, levels of IGF2 DMR0 methylation are associated with increased levels of IGF2 protein and increased birth weight.Citation56 This relationship may help to explain the previously described links between non-central cord insertion, low placenta and birth weight and increased risk of neonatal morbidity and mortality.Citation48-Citation51 We suggest that future studies of twins and singletons should consider cord placement and placenta size as potential confounders. Furthermore, because of the link between low birth weight and risk for complex disease,Citation57 it will be important to look at the relationship between unique environmental factors and future health in studies of twins and singletons, and not just on factors common to both twins.
Gestational diabetes influences neonatal DNA methylation
A second striking finding of our study is the observed consistent negative association between maternal gestational diabetes and DNA methylation levels at IGF2/H19 in non-placental tissues ( and ; Table S2). Studies of rodents have investigated such a link and both found a positive association between induced gestational diabetes and DNA methylation at the IGF2/H19 ICRCitation58,Citation59 and IGF2 DMR2.Citation59 The discrepancy between these findings and our study could be due to the differences in the control of imprinting between species or to the multivariable nature of our study, which takes account of the potential effects of a range of factors acting simultaneously.
The complexity of IGF2/H19 methylation differences across tissue and loci associated with different maternal factors
We have reported associations between early life environment and DNA methylation in four DMRs of the IGF2/H19 locus in multiple tissues at birth. This is the first study that has investigated multiple early environmental factors across multiple tissues and loci. Overall, we observed a complex relationship with different magnitudes and directions of effect on DNA methylation variation in association with varying gestational factors across five tissue types ( and –; Table S2). The exception is the position of umbilical cord insertion, specifically in MC twins, that showed a consistent direction of effect in all tissues. Although it has been shown that mammalian tissues can differ in their epigenetic status (e.g. refs. Citation46,Citation60), there have been few studies of the effects of early environment on DNA methylation in different tissues. However, our findings agreed with earlier data from studies of mature mice and humans (reviewed in ref. Citation61) and with recent genome-scale studies: in mice of early alcohol exposureCitation62 and in humans of assisted reproductive technologies ART,Citation63 smoking,Citation35 maternal homocysteine levels,Citation64 and childhood stress.Citation65 Taken together, these data showed that the effects of early environment on DNA methylation in offspring are often locus-specific and therefore effects from other environmental factors (or even the same factors) cannot be assumed to be the same for other genes or tissues. Our study also provides the first insights into the potential processes underlying the variable tissue-specific effects on genome-scale DNA methylation that we have recently described at birth.Citation45
Comparison of effect direction with previously published data
Our DNA methylation data from multiple maternal factors, cell types and IGF2/H19 DMRs allow comparisons with previous studies in rodents and humans (). Although the effect sizes might be small, most of the top hits in epigenome-wide epigenetic studies are only around 5% and, when occurring in gene networks, may be additiveCitation66 and should be considered as relative rather than absolute differences.Citation67 A recent study showed that an IGF2/H19 methylation difference of 5% was associated with a 2 mm difference in abdominal skin fold thickness in children age 1–10 y and that a 3.4% IGF2/H19 methylation difference was associated with a 18 mm difference in their head circumferences.Citation68 This shows that small differences in methylation can be associated with sizable phenotypic effects. Furthermore, relationships between DNA methylation and gene expression have been demonstrated to be nonlinear, with small changes in DNA methylation being associated with large differences in gene expression.Citation20,Citation69-Citation71
Table 3. Comparisons with previously published data
With reference to , previous data showing a positive correlation between maternal periconceptional folic acid supplementation and DNA methylation at the IGF2 DMR0 in whole blood from 17 mo-old offspring in humansCitation40 agree with our positive, although non-significant, association between the same two variables in granulocyte and CBMC blood fractions taken at birth. Conversely, previous dataCitation39 showed a negative correlation between maternal folic acid supplementation during pregnancy and DNA methylation at the first CTCF binding site within the IGF2/H19 ICR in whole blood from newborn mouse pups, which disagrees with our positive, although non-significant, association between the same exposure-DMR combination in blood fractions taken at birth. Differences between the locations of the assayed regions within the IGF2/H19 ICR could explain the discrepancy. Few studies have looked at the effects of prenatal alcohol exposure on IGF2/H19 DNA methylation. Animal studies have shown a trend of decreased IGF2/H19 ICR methylation in placenta,Citation28 which was comparable to our study, as reduction of placental IGF2/H19 ICR methylation was observed in mothers who consumed alcohol during the periconceptional period.
DNA methylation at the IGF2 DMR0 was reduced by 5.2%Citation37 and 2.1%Citation72 and reduced by 0.5% in the H19 promoter DMRCitation72 in whole blood of 60-y-olds whose mothers were exposed to famine during early gestation compared with their unexposed siblings. This data can be compared with our variable of maternal macronutrient intake, as this would be mainly compromised in famine conditions. Indeed, the Dutch famine data agree with our findings of a 1.0 to 1.7% (non-significant) increase in DNA methylation at the IGF2 DMR0 and a 0.5% (non-significant) increase in DNA methylation at the H19 promoter DMR in CBMC and granulocyte blood fractions for every 1 standard deviation increase in macronutrient score ( and ). Finally, a positive association between maternal periconceptional smoking and DNA methylation at the IGF2 DMR0 in whole blood at birth (significant in males onlyCitation36) agrees with our data of a positive, but non-significant association between DNA methylation at the IGF2 DMR0 in neonatal blood fractions ( and ).
We focused on the IGF2 locus because IGF2 and H19 the genes within are highly expressed in utero where they contribute to fetal and placental growth.Citation15 However, other genes involved in fetal growth, imprinted or otherwise, will need to be examined in future studies to determine how widespread these effects are.
Conclusions
For the first time we present evidence for the effect of specific non-shared environments and for locus-specific effects of a number of shared environments on the neonatal epigenome. Our study has a number of strengths including the use of multiple cell types, multiple DMRs within the IGF2/H19 locus and the rich data on maternal and supply line factors collected. In addition, our data underwent stringent QC analysis and excluded CpGs previously shown to be under genetic influence.Citation45 We also present evidence that in some cases, effects of a particular environment may be locus- or tissue-specific. Further studies will need to be conducted to determine the effect of the small effect sizes we found (1.6% to 3.0% absolute differences for significant associations in all tissues/DMRs combined [] and up to 8.7% for absolute difference for assay mean/tissue associations [‒]), on levels of the corresponding genes and proteins. Other studies have shown such effects to be nonlinearCitation20,Citation69,Citation70 and it has been suggested that such effects at multiple genes may be additive.Citation66 Further studies are also needed to determine whether such small differences are associated with disease risk and whether they are stable over time. If these and other epigenetic differences at birth are found to confer such risk, then for shared factors, this could be addressed by targeted intervention during pregnancy.Citation73 As intervention is usually not possible with supply line factors, they could at least be closely monitored during pregnancy.Citation73 In the future, risk-associated gene- or pathway-specific risks effects could be reversed by dietary, or pharmaceutical or other interventions.Citation74 Indeed, such broad-based interventions have already proven successful at reversing the risks associated with fetal programming in animal models.Citation75 Our study has shown that the epigenetic effects of such interventions will need to be studied in multiple tissues.
Materials and Methods
Twins cohort
A subgroup of 67 monozygotic (MZ) and 49 dizygotic (DZ) twin pairs from the PETS cohortCitation52 were included in this study. Questionnaire data were obtained for maternal nutritional intake, lifestyle and presence of gestational diabetes at different time points of pregnancy (see below for details). The majority of the twin pairs were same sex with only 9.5% being of opposite sex within a pair. An overview of the sample characteristics and maternal data used for thus study is listed in . The study was performed with appropriate human ethics clearances from the Royal Women’s Hospital (06/21), Mercy Hospital for Women (R06/30), and Monash Medical Centre (06117C), Melbourne.
Maternal and supply line factors
Data on maternal and supply line factors were collected as previously described.Citation52 Location of the umbilical cord insertion into the placenta was measured at birth and defined either as “central” (located in the inner 50% of the placental radius) or “non-central” (located with peripheral or velamentous positioning.).Citation50 Placental weight was measured at birth, with total placenta weight being shared equally between co-twins when values for separate placentas were unavailable (e.g., in MC twin pairs). Folate intake from nutritional supplements was obtained during the first 12 weeks of pregnancy (yes/no) and serum vitamin B12 and homocysteine levels measured from a sample of peripheral blood taken from mothers at 28 weeks’ gestation. Protein, carbohydrate and energy intake in the first two trimesters of pregnancy were obtained through a validated food frequency questionnaireCitation76 at a median gestational age of 28 weeks. Alcohol consumption and smoking before the mother knew she was pregnant (yes/no) were obtained from mothers between 19 and 24 weeks of gestation. Maternal perceived stress score was obtained via questionnaireCitation77 at 28 weeks gestation and the presence of gestational diabetes was ascertained by questionnaires at recruitment, 24 weeks and, where appropriate, 36 weeks’ gestation.
Tissue collection and cell processing
Placenta, umbilical cords, cord blood and buccals were collected at the time of delivery and processed soon after; CBMCs and granulocytes were isolated from cord blood, and HUVECs from cords as previously described.Citation45
DNA extraction, bisulphite conversion, and locus-specific methylation analysis
DNA was prepared from buccal cells via salt extraction and from all other tissues by standard phenol:chloroform extraction and ethanol precipitation as described previously.Citation45 A total of 500 ng to 1 µg genomic DNA was bisulphite converted using the MethylEasy Exceed Rapid Bisulphite Modification Kit (Human Genetic Signatures) and IGF2/H19 DMRs amplified as described previously, named according to location.Citation45 The two in the promoter regions of IGF2 and H19 are referred to as IGF2 DMR0 (chr11:2,169,514-chr11:2,169,771) and H19 promoter DMR (chr11:2,019,654-chr11:2,019,849) respectively (; Table S1). IGF2/H19 ICR (chr11:2,021,010-chr11:2,021,266) is located within the 6th CTCF site of the ICR, and IGF2 DMR2 (chr11:2,154,349-chr11:2,154,067) is located within exon 9 of IGF2 (; Table S1). DNA methylation was measured using the MassARRAY EpiTYPER (SEQUENOM Inc.). At least two replicate amplifications were performed in all instances and data was subjected to stringent cleaning steps as outlined previously.Citation45 Study individuals were genotyped for all known SNPs in the regions analyzed and where a SNP was found to abolish a CpG site in one or both alleles, methylation values for this CpG site or CpG unit containing the SNP were set to missing.
Statistical analysis
Multiple linear regression of methylation data jointly on the environmental measures was used to assess associations, with robust standard errors used to account for correlation of methylation data within the same subject, and to allow for potential correlation between co-twin methylation values. This approach does not impose any particular covariance structure on the data. The stability of the covariance estimation was assessed by checking the results for consistency with those obtained from a hierarchical random-effects model. In the latter model, loci were clustered within tissue, tissues were clustered within baby and twins were clustered within mother. An order one autoregressive (AR1) covariance structure for correlation of CpG units within each locus was imposed, reflecting generally positive correlations within a locus that decay with distance. We chose to report results from the model with unstructured covariance, as these are more robust to misspecification of the covariance structure. Since it is unknown whether the associations, if any, should hold generally across all four loci and five cell types at specific loci only, or only for specific tissues, in addition to reporting overall associations pooled across all loci and tissue types, interactions by locus and by tissue were fitted to explore differences in effects across loci and across tissues. Effect sizes were also explored separately for each combination of locus and tissue, but at this level of detail, formal testing of interactions was not considered feasible. DNA methylation levels were converted to z-scores prior to analysis to make them comparable across CpG units and tissues. High correlations within the set of macronutrient/energy measures were handled by using a principle components analysis to derive summary variables. The first principal component, hereafter referred to simply as macronutrient score, was chosen for use in analyses as it captured 93% of the variation between protein, carbohydrate and energy and represented an approximate equal weighting of these three measures. To facilitate comparison of effect sizes across all environmental measures, continuous measures were converted to z-scores prior to analysis. For reporting, regression coefficients were converted to percentage difference in mean methylation by multiplying each regression coefficient by the average standard deviation of methylation across CpG units. The potential for false positives from multiple testing was addressed by limiting the number of regression coefficients estimated to four groups of ten (one group of ten factors per locus). Formal control of the false positive rate is not possible in this setting, however, due to the moderate number of potentially correlated comparisons being performed. All analyses were performed using Stata statistical software (StataCorp.).
| Abbreviation: | ||
| MZ | = | monozygotic |
| DZ | = | dizygotic |
| MC | = | monochorionic |
| DC | = | dichorionic |
| IGF2 | = | Insulin-like Growth Factor 2 |
| DMR | = | differentially methylated region |
| ICR | = | imprinting control region |
| CTCF | = | CCCTC-binding factor |
| HUVECs | = | human umbilical vein endothelial cells |
| CBMCs | = | cord blood mononuclear cells |
| PETS | = | peri/postnatal epigenetic twins study |
Additional material
Download Zip (148.7 KB)Acknowledgments
We wish to thank John Carlin, Clinical Epidemiology and Biostatistics Unit, MCRI, Mark Umstad, Royal Women’s Hospital, Melbourne, Euan Wallace, Monash Medical Centre, Melbourne and Mark Permezel, Mercy Hospital for Women, Melbourne for their contributions to establishing the PETS cohort; Sarah Healy, Tin Vaiano, Nicole Brookes, Jennifer Foord, Sheila Holland, Anne Krastev, Siva Illancheran and Joanne Mockler for recruitment and sample collection; Technical officer Anna Czajko, Study Coordinator Geraldine McIlroy, and all mothers and twins that participated in this study.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Financial Disclosures
This work was supported by grants from the Australian National Health and Medical Research Council [grant numbers 437015, 607358 to JMC and RS]; the Financial Markets Foundation for Children (grant number 032-2007); and the Victorian Government’s Operational Infrastructure Support Program. JMC and YJL would like to acknowledge financial support from the Murdoch Childrens Research Institute. The authors have no competing financial interests.
Supplemental Materials
Supplemental materials may be found here: http://www.landesbioscience.com/journals/epigenetics/article/25908
References
- Gibney ER, Nolan CM. Epigenetics and gene expression. Heredity (Edinb) 2010; 105:4 - 13; http://dx.doi.org/10.1038/hdy.2010.54; PMID: 20461105
- Vaissière T, Sawan C, Herceg Z. Epigenetic interplay between histone modifications and DNA methylation in gene silencing. Mutat Res 2008; 659:40 - 8; http://dx.doi.org/10.1016/j.mrrev.2008.02.004; PMID: 18407786
- Numata S, Ye TZ, Hyde TM, Guitart-Navarro X, Tao R, Wininger M, et al. DNA methylation signatures in development and aging of the human prefrontal cortex. Am J Hum Genet 2012; 90:260 - 72; http://dx.doi.org/10.1016/j.ajhg.2011.12.020; PMID: 22305529
- Gluckman PD, Hanson MA, Buklijas T, Low FM, Beedle AS. Epigenetic mechanisms that underpin metabolic and cardiovascular diseases. Nat Rev Endocrinol 2009; 5:401 - 8; http://dx.doi.org/10.1038/nrendo.2009.102; PMID: 19488075
- Barker DJ, Hales CN, Fall CH, Osmond C, Phipps K, Clark PM. Type 2 (non-insulin-dependent) diabetes mellitus, hypertension and hyperlipidaemia (syndrome X): relation to reduced fetal growth. Diabetologia 1993; 36:62 - 7; http://dx.doi.org/10.1007/BF00399095; PMID: 8436255
- Hales CN, Barker DJ. Type 2 (non-insulin-dependent) diabetes mellitus: the thrifty phenotype hypothesis. Diabetologia 1992; 35:595 - 601; http://dx.doi.org/10.1007/BF00400248; PMID: 1644236
- Burdge GC, Slater-Jefferies J, Torrens C, Phillips ES, Hanson MA, Lillycrop KA. Dietary protein restriction of pregnant rats in the F0 generation induces altered methylation of hepatic gene promoters in the adult male offspring in the F1 and F2 generations. Br J Nutr 2007; 97:435 - 9; http://dx.doi.org/10.1017/S0007114507352392; PMID: 17313703
- Gluckman PD, Hanson MA, Cooper C, Thornburg KL. Effect of in utero and early-life conditions on adult health and disease. N Engl J Med 2008; 359:61 - 73; http://dx.doi.org/10.1056/NEJMra0708473; PMID: 18596274
- Reik W, Dean W, Walter J. Epigenetic reprogramming in mammalian development. Science 2001; 293:1089 - 93; http://dx.doi.org/10.1126/science.1063443; PMID: 11498579
- Skinner MK. Role of epigenetics in developmental biology and transgenerational inheritance. Birth Defects Res C Embryo Today 2011; 93:51 - 5; http://dx.doi.org/10.1002/bdrc.20199; PMID: 21425441
- Constância M, Pickard B, Kelsey G, Reik W. Imprinting mechanisms. Genome Res 1998; 8:881 - 900; PMID: 9750189
- Woodfine K, Huddleston JE, Murrell A. Quantitative analysis of DNA methylation at all human imprinted regions reveals preservation of epigenetic stability in adult somatic tissue. Epigenetics Chromatin 2011; 4:1; http://dx.doi.org/10.1186/1756-8935-4-1; PMID: 21281512
- Li Y, Sasaki H. Genomic imprinting in mammals: its life cycle, molecular mechanisms and reprogramming. Cell Res 2011; 21:466 - 73; http://dx.doi.org/10.1038/cr.2011.15; PMID: 21283132
- Bartolomei MS. Genomic imprinting: employing and avoiding epigenetic processes. Genes Dev 2009; 23:2124 - 33; http://dx.doi.org/10.1101/gad.1841409; PMID: 19759261
- Gabory A, Jammes H, Dandolo L. The H19 locus: role of an imprinted non-coding RNA in growth and development. Bioessays 2010; 32:473 - 80; http://dx.doi.org/10.1002/bies.200900170; PMID: 20486133
- Takai D, Gonzales FA, Tsai YC, Thayer MJ, Jones PA. Large scale mapping of methylcytosines in CTCF-binding sites in the human H19 promoter and aberrant hypomethylation in human bladder cancer. Hum Mol Genet 2001; 10:2619 - 26; http://dx.doi.org/10.1093/hmg/10.23.2619; PMID: 11726548
- Guo L, Choufani S, Ferreira J, Smith A, Chitayat D, Shuman C, et al. Altered gene expression and methylation of the human chromosome 11 imprinted region in small for gestational age (SGA) placentae. Dev Biol 2008; 320:79 - 91; http://dx.doi.org/10.1016/j.ydbio.2008.04.025; PMID: 18550048
- Murrell A, Ito Y, Verde G, Huddleston J, Woodfine K, Silengo MC, et al. Distinct methylation changes at the IGF2-H19 locus in congenital growth disorders and cancer. PLoS One 2008; 3:e1849; http://dx.doi.org/10.1371/journal.pone.0001849; PMID: 18365005
- Sullivan MJ, Taniguchi T, Jhee A, Kerr N, Reeve AE. Relaxation of IGF2 imprinting in Wilms tumours associated with specific changes in IGF2 methylation. Oncogene 1999; 18:7527 - 34; http://dx.doi.org/10.1038/sj.onc.1203096; PMID: 10602511
- Lillycrop KA, Phillips ES, Jackson AA, Hanson MA, Burdge GC. Dietary protein restriction of pregnant rats induces and folic acid supplementation prevents epigenetic modification of hepatic gene expression in the offspring. J Nutr 2005; 135:1382 - 6; PMID: 15930441
- Lillycrop KA, Slater-Jefferies JL, Hanson MA, Godfrey KM, Jackson AA, Burdge GC. Induction of altered epigenetic regulation of the hepatic glucocorticoid receptor in the offspring of rats fed a protein-restricted diet during pregnancy suggests that reduced DNA methyltransferase-1 expression is involved in impaired DNA methylation and changes in histone modifications. Br J Nutr 2007; 97:1064 - 73; http://dx.doi.org/10.1017/S000711450769196X; PMID: 17433129
- Waterland RA, Jirtle RL. Transposable elements: targets for early nutritional effects on epigenetic gene regulation. Mol Cell Biol 2003; 23:5293 - 300; http://dx.doi.org/10.1128/MCB.23.15.5293-5300.2003; PMID: 12861015
- Wolff GL, Kodell RL, Moore SR, Cooney CA. Maternal epigenetics and methyl supplements affect agouti gene expression in Avy/a mice. FASEB J 1998; 12:949 - 57; PMID: 9707167
- Waterland RA, Dolinoy DC, Lin JR, Smith CA, Shi X, Tahiliani KG. Maternal methyl supplements increase offspring DNA methylation at Axin Fused. Genesis 2006; 44:401 - 6; http://dx.doi.org/10.1002/dvg.20230; PMID: 16868943
- Saravanan P. Role of maternal vitamin B12 on the metabolic health of the offspring: a contributor to the diabetes epidemic?. Br J Diabetes Vasc Dis 2010; 10:109 - 14; http://dx.doi.org/10.1177/1474651409358015
- Yajnik CS, Deshpande SS, Jackson AA, Refsum H, Rao S, Fisher DJ, et al. Vitamin B12 and folate concentrations during pregnancy and insulin resistance in the offspring: the Pune Maternal Nutrition Study. Diabetologia 2008; 51:29 - 38; http://dx.doi.org/10.1007/s00125-007-0793-y; PMID: 17851649
- Sittig LJ, Redei EE. Paternal genetic contribution influences fetal vulnerability to maternal alcohol consumption in a rat model of fetal alcohol spectrum disorder. PLoS One 2010; 5:e10058; http://dx.doi.org/10.1371/journal.pone.0010058; PMID: 20383339
- Haycock PC, Ramsay M. Exposure of mouse embryos to ethanol during preimplantation development: effect on DNA methylation in the h19 imprinting control region. Biol Reprod 2009; 81:618 - 27; http://dx.doi.org/10.1095/biolreprod.108.074682; PMID: 19279321
- Stouder C, Somm E, Paoloni-Giacobino A. Prenatal exposure to ethanol: a specific effect on the H19 gene in sperm. Reprod Toxicol 2011; 31:507 - 12; http://dx.doi.org/10.1016/j.reprotox.2011.02.009; PMID: 21382472
- Darnaudéry M, Maccari S. Epigenetic programming of the stress response in male and female rats by prenatal restraint stress. Brain Res Rev 2008; 57:571 - 85; http://dx.doi.org/10.1016/j.brainresrev.2007.11.004; PMID: 18164765
- Liu D, Diorio J, Tannenbaum B, Caldji C, Francis D, Freedman A, et al. Maternal care, hippocampal glucocorticoid receptors, and hypothalamic-pituitary-adrenal responses to stress. Science 1997; 277:1659 - 62; http://dx.doi.org/10.1126/science.277.5332.1659; PMID: 9287218
- Oberlander TF, Weinberg J, Papsdorf M, Grunau R, Misri S, Devlin AM. Prenatal exposure to maternal depression, neonatal methylation of human glucocorticoid receptor gene (NR3C1) and infant cortisol stress responses. Epigenetics 2008; 3:97 - 106; http://dx.doi.org/10.4161/epi.3.2.6034; PMID: 18536531
- Liu Y, Murphy SK, Murtha AP, Fuemmeler BF, Schildkraut J, Huang Z, et al. Depression in pregnancy, infant birth weight and DNA methylation of imprint regulatory elements. Epigenetics 2012; 7:735 - 46; http://dx.doi.org/10.4161/epi.20734; PMID: 22677950
- Cooper WN, Khulan B, Owens S, Elks CE, Seidel V, Prentice AM, et al. DNA methylation profiling at imprinted loci after periconceptional micronutrient supplementation in humans: results of a pilot randomized controlled trial. FASEB J 2012; 26:1782 - 90; http://dx.doi.org/10.1096/fj.11-192708; PMID: 22267336
- Breton CV, Byun HM, Wenten M, Pan F, Yang A, Gilliland FD. Prenatal tobacco smoke exposure affects global and gene-specific DNA methylation. Am J Respir Crit Care Med 2009; 180:462 - 7; http://dx.doi.org/10.1164/rccm.200901-0135OC; PMID: 19498054
- Murphy SK, Adigun A, Huang Z, Overcash F, Wang F, Jirtle RL, et al. Gender-specific methylation differences in relation to prenatal exposure to cigarette smoke. Gene 2012; 494:36 - 43; http://dx.doi.org/10.1016/j.gene.2011.11.062; PMID: 22202639
- Heijmans BT, Tobi EW, Stein AD, Putter H, Blauw GJ, Susser ES, et al. Persistent epigenetic differences associated with prenatal exposure to famine in humans. Proc Natl Acad Sci U S A 2008; 105:17046 - 9; http://dx.doi.org/10.1073/pnas.0806560105; PMID: 18955703
- Waterland RA, Kellermayer R, Laritsky E, Rayco-Solon P, Harris RA, Travisano M, et al. Season of conception in rural gambia affects DNA methylation at putative human metastable epialleles. PLoS Genet 2010; 6:e1001252; http://dx.doi.org/10.1371/journal.pgen.1001252; PMID: 21203497
- Hoyo C, Murtha AP, Schildkraut JM, Jirtle RL, Demark-Wahnefried W, Forman MR, et al. Methylation variation at IGF2 differentially methylated regions and maternal folic acid use before and during pregnancy. Epigenetics 2011; 6:928 - 36; http://dx.doi.org/10.4161/epi.6.7.16263; PMID: 21636975
- Steegers-Theunissen RP, Obermann-Borst SA, Kremer D, Lindemans J, Siebel C, Steegers EA, et al. Periconceptional maternal folic acid use of 400 microg per day is related to increased methylation of the IGF2 gene in the very young child. PLoS One 2009; 4:e7845; http://dx.doi.org/10.1371/journal.pone.0007845; PMID: 19924280
- Ba Y, Yu H, Liu F, Geng X, Zhu C, Zhu Q, et al. Relationship of folate, vitamin B12 and methylation of insulin-like growth factor-II in maternal and cord blood. Eur J Clin Nutr 2011; 65:480 - 5; http://dx.doi.org/10.1038/ejcn.2010.294; PMID: 21245875
- Lopes MC, Andrew T, Carbonaro F, Spector TD, Hammond CJ. Estimating heritability and shared environmental effects for refractive error in twin and family studies. Invest Ophthalmol Vis Sci 2009; 50:126 - 31; http://dx.doi.org/10.1167/iovs.08-2385; PMID: 18757506
- Heijmans BT, Kremer D, Tobi EW, Boomsma DI, Slagboom PE. Heritable rather than age-related environmental and stochastic factors dominate variation in DNA methylation of the human IGF2/H19 locus. Hum Mol Genet 2007; 16:547 - 54; http://dx.doi.org/10.1093/hmg/ddm010; PMID: 17339271
- Wong CC, Caspi A, Williams B, Craig IW, Houts R, Ambler A, et al. A longitudinal study of epigenetic variation in twins. Epigenetics 2010; 5:516 - 26; http://dx.doi.org/10.4161/epi.5.6.12226; PMID: 20505345
- Ollikainen M, Smith KR, Joo EJ, Ng HK, Andronikos R, Novakovic B, et al. DNA methylation analysis of multiple tissues from newborn twins reveals both genetic and intrauterine components to variation in the human neonatal epigenome. Hum Mol Genet 2010; 19:4176 - 88; http://dx.doi.org/10.1093/hmg/ddq336; PMID: 20699328
- Gordon L, Joo JE, Powell JE, Ollikainen M, Novakovic B, Li X, et al. Neonatal DNA methylation profile in human twins is specified by a complex interplay between intrauterine environmental and genetic factors, subject to tissue-specific influence. Genome Res 2012; 22:1395 - 406; http://dx.doi.org/10.1101/gr.136598.111; PMID: 22800725
- Machin GA. Some causes of genotypic and phenotypic discordance in monozygotic twin pairs. Am J Med Genet 1996; 61:216 - 28; http://dx.doi.org/10.1002/(SICI)1096-8628(19960122)61:3<216::AID-AJMG5>3.0.CO;2-S; PMID: 8741866
- Lopriore E, Pasman SA, Klumper FJ, Middeldorp JM, Walther FJ, Oepkes D. Placental characteristics in growth-discordant monochorionic twins: a matched case-control study. Placenta 2012; 33:171 - 4; http://dx.doi.org/10.1016/j.placenta.2011.12.004; PMID: 22197628
- De Paepe ME, Shapiro S, Hanley LC, Chu S, Luks FI. Correlation between cord insertion type and superficial choriovasculature in diamniotic-monochorionic twin placentas. Placenta 2011; 32:901 - 5; http://dx.doi.org/10.1016/j.placenta.2011.08.001; PMID: 21864904
- Kent EM, Breathnach FM, Gillan JE, McAuliffe FM, Geary MP, Daly S, et al. Placental cord insertion and birthweight discordance in twin pregnancies: results of the national prospective ESPRiT Study. Am J Obstet Gynecol 2011; 205:e1 - 7; http://dx.doi.org/10.1016/j.ajog.2011.06.077; PMID: 21864823
- Hack KE, Nikkels PG, Koopman-Esseboom C, Derks JB, Elias SG, van Gemert MJ, et al. Placental characteristics of monochorionic diamniotic twin pregnancies in relation to perinatal outcome. Placenta 2008; 29:976 - 81; http://dx.doi.org/10.1016/j.placenta.2008.08.019; PMID: 18835495
- Saffery R, Morley R, Carlin JB, Joo JH, Ollikainen M, Novakovic B, et al. Cohort profile: The peri/post-natal epigenetic twins study. Int J Epidemiol 2012; 41:55 - 61; http://dx.doi.org/10.1093/ije/dyr140; PMID: 22422448
- Tycko B. Imprinted genes in placental growth and obstetric disorders. Cytogenet Genome Res 2006; 113:271 - 8; http://dx.doi.org/10.1159/000090842; PMID: 16575190
- Jones CT, Parer JT. The effect of alterations in placental blood flow on the growth of and nutrient supply to the fetal guinea-pig. J Physiol 1983; 343:525 - 37; PMID: 6358463
- Yampolsky M, Salafia CM, Shlakhter O, Haas D, Eucker B, Thorp J. Centrality of the umbilical cord insertion in a human placenta influences the placental efficiency. Placenta 2009; 30:1058 - 64; http://dx.doi.org/10.1016/j.placenta.2009.10.001; PMID: 19879649
- Hoyo C, Fortner K, Murtha AP, Schildkraut JM, Soubry A, Demark-Wahnefried W, et al. Association of cord blood methylation fractions at imprinted insulin-like growth factor 2 (IGF2), plasma IGF2, and birth weight. Cancer Causes Control 2012; 23:635 - 45; http://dx.doi.org/10.1007/s10552-012-9932-y; PMID: 22392079
- Barker DJ. The fetal and infant origins of adult disease. BMJ 1990; 301:1111; http://dx.doi.org/10.1136/bmj.301.6761.1111; PMID: 2252919
- Shao WJ, Tao LY, Gao C, Xie JY, Zhao RQ. Alterations in methylation and expression levels of imprinted genes H19 and Igf2 in the fetuses of diabetic mice. Comp Med 2008; 58:341 - 6; PMID: 18724775
- Ding GL, Wang FF, Shu J, Tian S, Jiang Y, Zhang D, et al. Transgenerational glucose intolerance with Igf2/H19 epigenetic alterations in mouse islet induced by intrauterine hyperglycemia. Diabetes 2012; 61:1133 - 42; http://dx.doi.org/10.2337/db11-1314; PMID: 22447856
- Rakyan VK, Down TA, Thorne NP, Flicek P, Kulesha E, Gräf S, et al. An integrated resource for genome-wide identification and analysis of human tissue-specific differentially methylated regions (tDMRs). Genome Res 2008; 18:1518 - 29; http://dx.doi.org/10.1101/gr.077479.108; PMID: 18577705
- Kim YI. Folate and DNA methylation: a mechanistic link between folate deficiency and colorectal cancer?. Cancer Epidemiol Biomarkers Prev 2004; 13:511 - 9; PMID: 15066913
- Liu Y, Balaraman Y, Wang G, Nephew KP, Zhou FC. Alcohol exposure alters DNA methylation profiles in mouse embryos at early neurulation. Epigenetics. Official Journal Of The DNA Methylation Society 2009; 4:500 - 11; http://dx.doi.org/10.4161/epi.4.7.9925
- Katari S, Turan N, Bibikova M, Erinle O, Chalian R, Foster M, et al. DNA methylation and gene expression differences in children conceived in vitro or in vivo. Hum Mol Genet 2009; 18:3769 - 78; http://dx.doi.org/10.1093/hmg/ddp319; PMID: 19605411
- Fryer AA, Nafee TM, Ismail KM, Carroll WD, Emes RD, Farrell WE. LINE-1 DNA methylation is inversely correlated with cord plasma homocysteine in man: a preliminary study. Epigenetics 2009; 4:394 - 8; http://dx.doi.org/10.4161/epi.4.6.9766; PMID: 19755846
- Essex MJ, Thomas Boyce W, Hertzman C, Lam LL, Armstrong JM, Neumann SM, et al. Epigenetic Vestiges of Early Developmental Adversity: Childhood Stress Exposure and DNA Methylation in Adolescence. Child Dev 2011; PMID: 21883162
- Heijmans BT, Mill J. Commentary: The seven plagues of epigenetic epidemiology. Int J Epidemiol 2012; 41:74 - 8; http://dx.doi.org/10.1093/ije/dyr225; PMID: 22269254
- Heijmans BT, Tobi EW, Lumey LH, Slagboom PE. The epigenome: archive of the prenatal environment. Epigenetics 2009; 4:526 - 31; http://dx.doi.org/10.4161/epi.4.8.10265; PMID: 19923908
- Huang RC, Galati JC, Burrows S, Beilin LJ, Li X, Pennell CE, et al. DNA methylation of the IGF2/H19 imprinting control region and adiposity distribution in young adults. Clin Epigenetics 2012; 4:21; http://dx.doi.org/10.1186/1868-7083-4-21; PMID: 23148549
- Hsieh CL. Dependence of transcriptional repression on CpG methylation density. Mol Cell Biol 1994; 14:5487 - 94; PMID: 7518564
- Boyes J, Bird A. Repression of genes by DNA methylation depends on CpG density and promoter strength: evidence for involvement of a methyl-CpG binding protein. EMBO J 1992; 11:327 - 33; PMID: 1310933
- Tremolizzo L, Carboni G, Ruzicka WB, Mitchell CP, Sugaya I, Tueting P, et al. An epigenetic mouse model for molecular and behavioral neuropathologies related to schizophrenia vulnerability. Proc Natl Acad Sci U S A 2002; 99:17095 - 100; http://dx.doi.org/10.1073/pnas.262658999; PMID: 12481028
- Tobi EW, Slagboom PE, van Dongen J, Kremer D, Stein AD, Putter H, et al. Prenatal famine and genetic variation are independently and additively associated with DNA methylation at regulatory loci within IGF2/H19. PLoS One 2012; 7:e37933; http://dx.doi.org/10.1371/journal.pone.0037933; PMID: 22666415
- Martínez JA, Cordero P, Campión J, Milagro FI. Interplay of early-life nutritional programming on obesity, inflammation and epigenetic outcomes. Proc Nutr Soc 2012; 71:276 - 83; http://dx.doi.org/10.1017/S0029665112000055; PMID: 22390978
- Gluckman PD, Hanson MA. Developmental plasticity and human disease: research directions. J Intern Med 2007; 261:461 - 71; http://dx.doi.org/10.1111/j.1365-2796.2007.01802.x; PMID: 17444885
- Wyrwoll CS, Mark PJ, Waddell BJ. Developmental programming of renal glucocorticoid sensitivity and the renin-angiotensin system. Hypertension 2007; 50:579 - 84; http://dx.doi.org/10.1161/HYPERTENSIONAHA.107.091603; PMID: 17664394
- Hodge A, Patterson AJ, Brown WJ, Ireland P, Giles G. The Anti Cancer Council of Victoria FFQ: relative validity of nutrient intakes compared with weighed food records in young to middle-aged women in a study of iron supplementation. Aust N Z J Public Health 2000; 24:576 - 83; http://dx.doi.org/10.1111/j.1467-842X.2000.tb00520.x; PMID: 11215004
- Cohen S, Kamarck T, Mermelstein R. A global measure of perceived stress. J Health Soc Behav 1983; 24:385 - 96; http://dx.doi.org/10.2307/2136404; PMID: 6668417